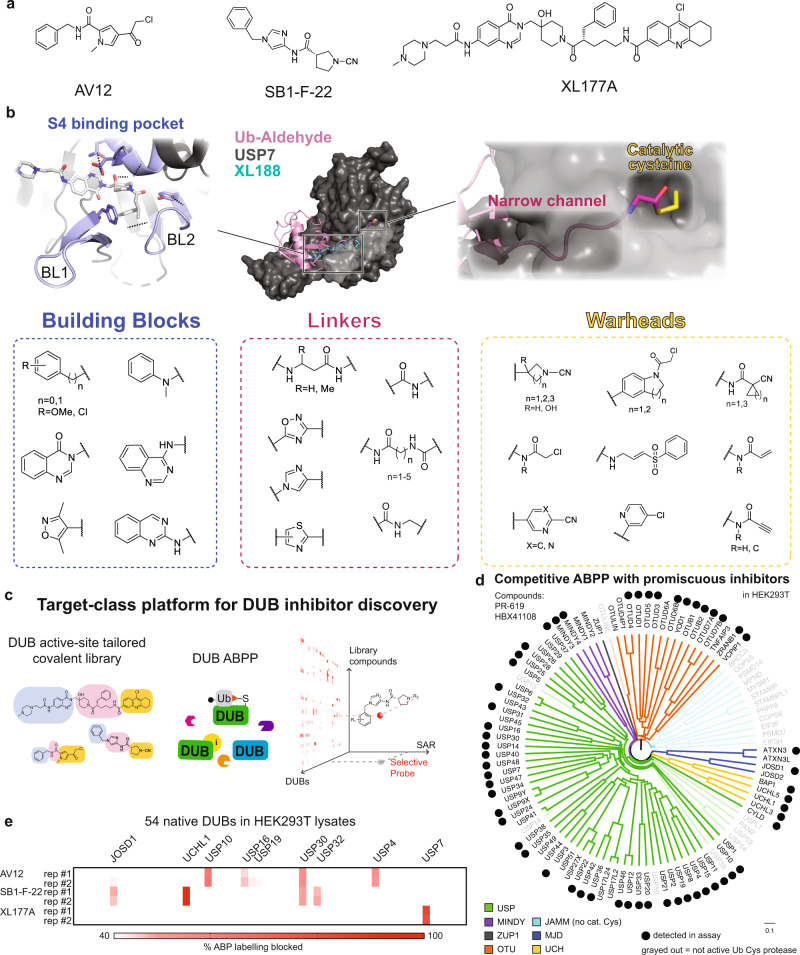
Fig. 1. A DUB-tailored inhibitor discovery platform.
a Structures of DUB inhibitors AV12, SB1-F-22, and XL177A. b Library compounds followed a three-piece modular design, where noncovalent building blocks, linkers, and warhead moieties were diversified to target conserved and divergent aspects of the DUB catalytic domain and nearby regions. Blue: Noncovalent building blocks were designed to target the blocking loops (BL) 1 and 2, in addition to the P4 site. Red: Linkers were designed to traverse a long and narrow channel leading up to the catalytic cysteine residue, occupied by the C-terminus of the substrate ubiquitin in catalysis. In the top panel, an overlay of crystal structures for USP7 with Ub-aldehyde (PDB: 1NBF) and USP7 with XL188 (PDB: 5VS6) are shown, with USP7 in gray, Ub-aldehyde in pink, and XL188 in cyan. Yellow: electrophilic warheads were designed to target the invariant catalytic cysteine residue shown in yellow in the top panel. c The target-class platform yields high content data to drive DUB hit identification and SAR for compound optimization. d Analysis of multitargeted covalent DUB inhibitors (HBX41108 and PR619) confirmed that our DUB-ABPP platform spanned members from all subfamilies of cysteine protease DUBs3,4. The dendrogram is colored by DUB subfamily. e ABPP analysis using covalent inhibitors depicted in (a) in duplicate runs (each shown individually) validated the ability of our platform to read-out selective compound binding activity with deep coverage of the human DUBome.