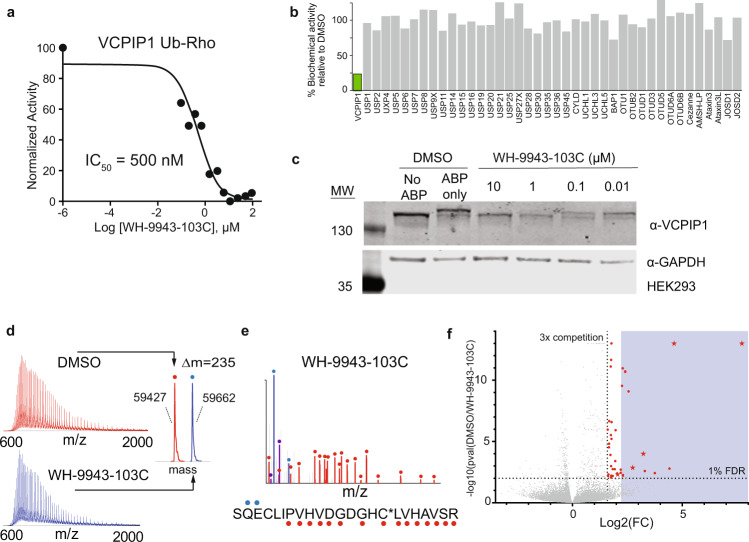
Fig. 5. VCPIP1 hit validation.
a Hit molecule WH-9943-103C inhibited deubiquitination activity of VCPIP1 in Ub-Rho cleavage assay after 6 h incubation (n = 1). b WH-9943-103C inhibited VCPIP1 (highlighted in green) selectively out of a panel of 41 purified recombinant DUBs after 15 min incubation. c WH-9943-103C displayed in-cell target engagement as determined for DUB labeling by ABP then visualized on a Western blot. d WH-9943-103C labeled recombinant VCPIP1 with 1:1 stoichiometry as read out by intact protein mass spectrometry. Labeled ion envelope is shown in blue, unlabeled ion envelope from DMSO control is shown in red. e CE-MS/MS identified the catalytic cysteine of VCPIP1 to be covalently modified by WH-9943-103C, red and blue glyphs adjacent to the peptide sequence indicate y- and b-type fragment ions, respectively, detected in the MS/MS spectrum. The y-ion series confirms catalytic cysteine modification (C*). f A proteome-wide competitive binding survey of WH-9943-103C activity (50µM) indicated off-target modification of 39 (red dots, FDR <1%, >3-fold competition relative to DMSO) out of 24,579 unique cysteines detected (p-values for all detected ratios derived from maximum likelihood estimation and corrected by the Benjamini-Hochberg method). The blue box indicates >4.5-fold competition relative to DMSO, the observed fold competition for VCPIP1. This includes 4 cysteines that were previously characterized as ‘hyper-reactive’ (red stars). Source data are provided as a source data file.