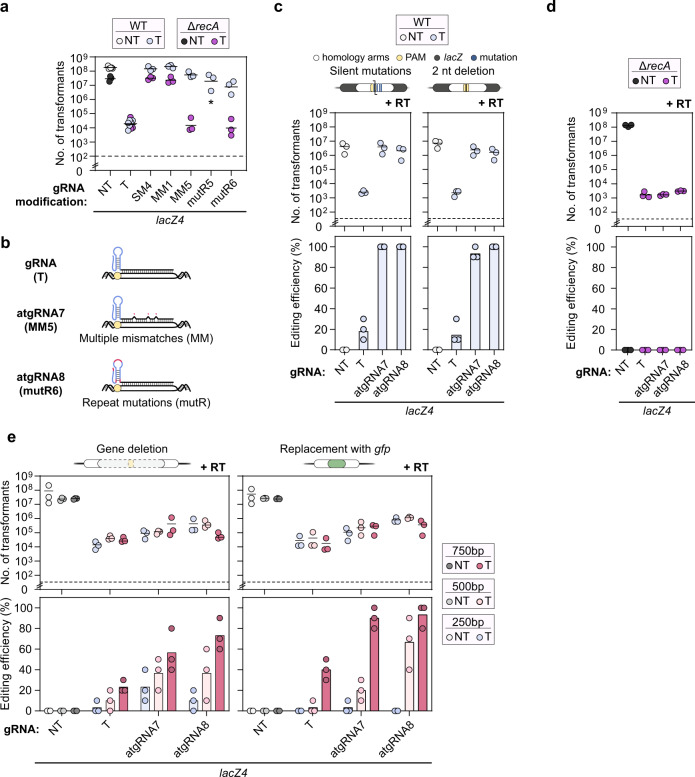
Fig. 3. CRISPR-driven editing with attenuated gRNAs extends to Cas12a.
a Genome targeting assay in E. coli and E. coli ΔrecA with modified gRNAs and Cas12a. b Schematic of atgRNAs for Cas12a. c) Genome editing assay in E. coli using atgRNAs to introduce a RE silent mutation (left panels) and a 2nt deletion in the lacZ ORF (right panels). d Editing assay to an AclI restriction enzyme site into lacZ in the E. coli ΔrecA strain. Each dot in the editing efficiency graph represents the value obtained for 3 colonies (n = 3). e Genome editing assay in E. coli using atgRNAs to delete the entire lacZ gene (left panels) and substitute lacZ with gfp (right panels). Individual dots for the transformations indicate a single biological replicate. NT indicates non-targeting and T indicates targeting. Individual dots for the editing efficiencies (in c–e) indicate the average of 3 colonies screened from one biological replicate for NT samples or 10 colonies screened from one biological replicate for targeting samples. Bars indicate the mean of the dots. * indicates that the transformants resulted in a lawn or uncountable colonies. The dashed line in (a, c, d) indicates the limit-of-detection. See Fig. 1 for details. WT indicates wild-type E. coli MG1655, ΔrecA indicates E. coli MG1655 ΔrecA, NT indicates non-targeting, T indicates targeting, and RT indicates repair template.