Fig. 4. An attenuated gRNA enhances editing in Klebsiella oxytoca.
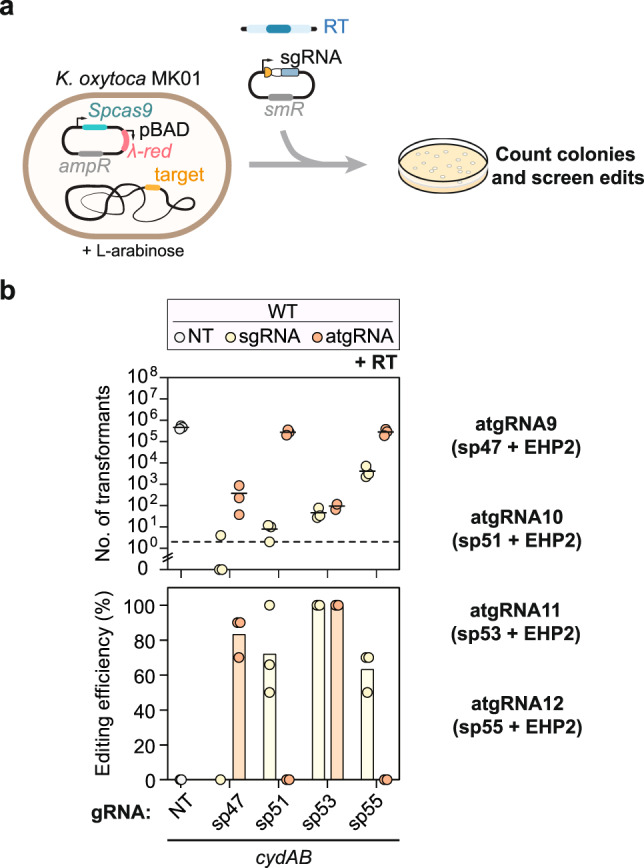
a Schematic of genome editing experimental setup. b Genome targeting assay in K. oxytoca with sgRNAs and atgRNAs to delete cydAB. Individual dots for the transformations indicate a single biological replicate. Dashed lines indicate the limit of detection from plating. NT indicates non-targeting. Individual dots for the editing efficiencies indicate the average of 3 colonies screened from one biological replicate for NT samples or 10 colonies screened from one biological replicate for targeting samples. Bars indicate the mean of the dots. The dashed line indicates the limit-of-detection. See Fig. 1 for details. The mean number of transformants (indicated by a horizontal line) was not calculated for samples with biological replicates that fell below the limit of detection. WT indicates wild-type K. oxytoca MK01, NT indicates non-targeting, and RT indicates repair template.
