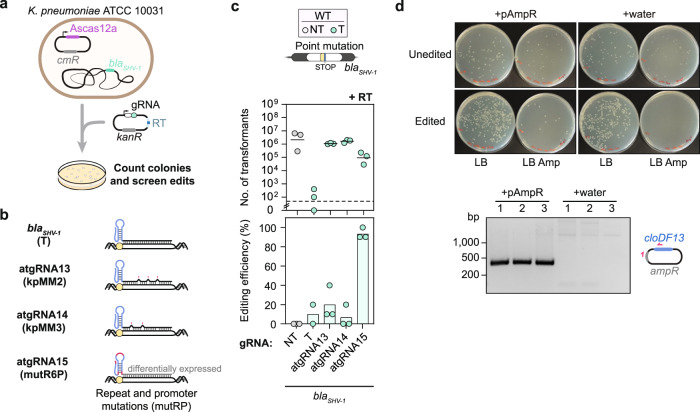
Fig. 5. Attenuated gRNAs can be applied to reverse antibiotic resistance in multidrug-resistant Klebsiella pneumoniae.
a Schematic of genome editing assay. b Schematic of canonical and attenuated guide design. c Genome editing assay in K. pneumoniae to introduce a stop codon in the blaSHV-1 gene. Each dot in the editing plot is the result obtained after screening 10 and 3 colonies respectively for the T and NT samples. Bars indicate the mean of the dots. For the T gRNA, each dot represents the editing efficiency obtained for 4 colonies due to the absence of other colonies on the plates. One of the biological replicates yielded no colonies for this condition. The dashed line indicates the limit-of-detection. See Fig. 1 for details. The mean number of transformants (indicated by a horizontal line) was not calculated for samples with biological replicates that fell below the limit-of-detection. d Top: plate images upon transformation of a single biological replicate of edited cells (atgRNA15) and unedited cells (NT) with the pAmpR plasmid or water in Amp or LB only plates. Bottom: schematic of pAmpR amplification by PCR with confirmatory gel image. The three lanes on the left are PCR amplicons from three individual colonies after plating the edited cells that were transformed with pAmpR, while the three lanes on the right are PCR amplicons from three individual colonies after plating edited cells that were transformed with water (from LB plates). The expected band size for the amplicon is ~400 bp. WT indicates wild-type K. pneumoniae ATCC 10031, NT indicates non-targeting, T indicates targeting, and RT indicates repair template.