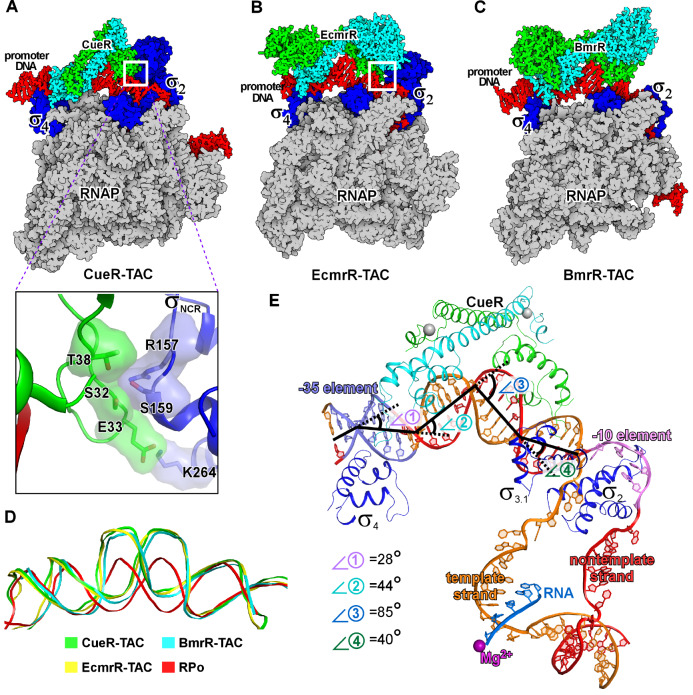
Figure5 .
The cryo-EM structures of transcription activation complexes comprising MerR-family TFsThe cryo-EM structures of (A) E. coli CueR transcription activation complex (PDB: 6XH7), (B) E. coli EcmrR transcription activation complex (PDB: 6XL5), and (C) B. subtilis BmrR transcription activation complex (PDB: 7CKQ). The insertion box shows the small surface patch of CueR-DBD that interacts with the σNCR. (D) Structure superimposition of the upstream dsDNAs of the above three transcription activation complexes and that of a bacterial RPo (PDB: 6OUL). (E) The kinks of the upstream promoter DNA at positions −35 (⦟1, 28°), −30 (⦟2, 44°), −24 (⦟3, 85°) and −18 (⦟4, 40°) in the cryo-EM structure of E. coli CueR-TAC (PDB: 6LDI). Kink 1 at −35 is induced by CueR and σ704, and kinks 2, 3 and 4 are induced by the CueR dimer.