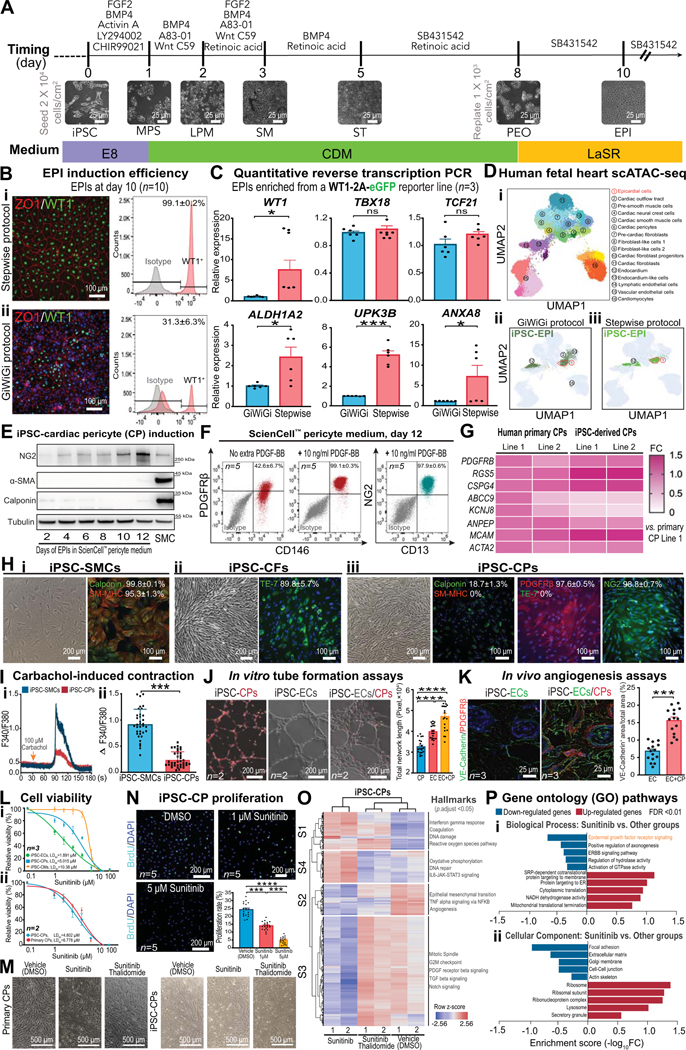
Figure. Phenotypic characterization and functional assessment of stepwise differentiated iPSC-cardiac pericytes.
A, A schematic showing stage-specific inhibition and activation of morphogens to generate pure epicardial cells (EPI) from human induced pluripotent stem cells (iPSCs). Representative bright-field images for each stage of cell differentiation are demonstrated. MPS, mid-primitive streak; LPM, lateral plate mesoderm; SM, splanchnic mesoderm; ST, septum transversum; PEO, pre-epicardial organ. B, Immunofluorescent images (ZO1 and WT1), flow cytometry (WT1) graphs (n=10 iPSC lines, 5M/5F), and quantitative data showing EPI induction efficiency by stepwise (i) and GiWiGi (ii) protocols. C, Quantitative reverse transcription PCR (RT-qPCR) results showing expression levels of canonical (WT1, TBX18, and TCF21) and mature (ALDH1A2, UPK3B, and ANXA8) markers of EPIs derived by both protocols. D, Single-cell ATAC sequencing (scATAC-seq) of EPIs generated by both GiWiGi (ii) and stepwise (iii) protocols are projected to that of human fetal heart cell clusters (i). Dotted frames indicate the EPI cluster in the human fetal heart scATAC-seq UMAP. E, Immunoblots showing time-dependent changes in cardiac pericytes (CP) and smooth muscle cell (SMC) markers during differentiation. iPSC-SMCs were used as a control. F, Flow cytometry and quantitative data (n=5, 2M/3F) showing iPSC-CP yields with and without exogenous platelet-derived growth factor (PDGF)-BB. G, A heatmap showing transcriptomic similarities of pericyte markers between primary and iPSC-CPs by RT-qPCR. H, Bright field and immunofluorescent images of iPSC-derived SMCs, cardiac fibroblasts (CFs), and endothelial cells (ECs). SM-MHC, smooth muscle-myosin heavy chain; NG2, neural/glial antigen 2. I, Calcium imaging using Fura-2 AM to quantitatively compare carbachol-induced intracellular calcium increases in iPSC-SMCs and iPSC-CPs. J, Tubular networks formed by iPSC-CPs, iPSC-ECs, and in combination in vitro (n=2, 1M/1F). iPSC-CPs were fluorescently labeled with Calcein Red-Orange AM. K, Vasculatures formed by iPSC-ECs alone (1.2×106 cells/Matrigel plug) or in combination with iPSC-CPs (1.0×106 iPSC-ECs and 0.2×106 iPSC-CPs/Matrigel plug) after being implanted in male immunodeficiency NOD SCID mice (n=3 per group) for 7 days. Antibodies targeting iPSC-ECs and iPSC-CPs are human-specific. L, Dose-response curves for (i) iPSC-CMs, iPSC-CPs, and iPSC-ECs (n=3, 1M/2F per group) and (ii) primary and iPSC-CPs (n=2, 1M/1F per group) after 72 hr of sunitinib (0 μM-100 μM) treatment using a PrestoBlue viability assay. M, Bright-filed images showing the cell morphology of primary and iPSC-CPs after 72 hr of vehicle, sunitinib (5 μM), and sunitinib (5 μM)/thalidomide (1 μM) treatment. N, BrdU incorporation assays showing suppressed iPSC-CP proliferation by sunitinib (n=5, 2M/3F per group) in a dose-dependent manner. O, A heatmap showing gene clustering patterns of iPSC-CPs treated with vehicle, sunitinib (5 μM), and sunitinib (5 μM)/thalidomide (1 μM) for 72 hr. Key changes of hallmarks in each cluster are highlighted. P, Gene ontology pathway analysis of iPSC-CPs treated with sunitinib versus other conditions. GraphPad Prism 9 was used for statistical analysis. All data are presented as mean ± sem. Statistical significance was assessed using a non-parametric statistical procedure (Wilcoxon signed-rank test for 2 groups and Kruskal-Wallis test for >2 groups). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns indicates not significant.