Figure 5. Patterned blocks of acidic and basic residues in IDRs are necessary and sufficient for selective partitioning.
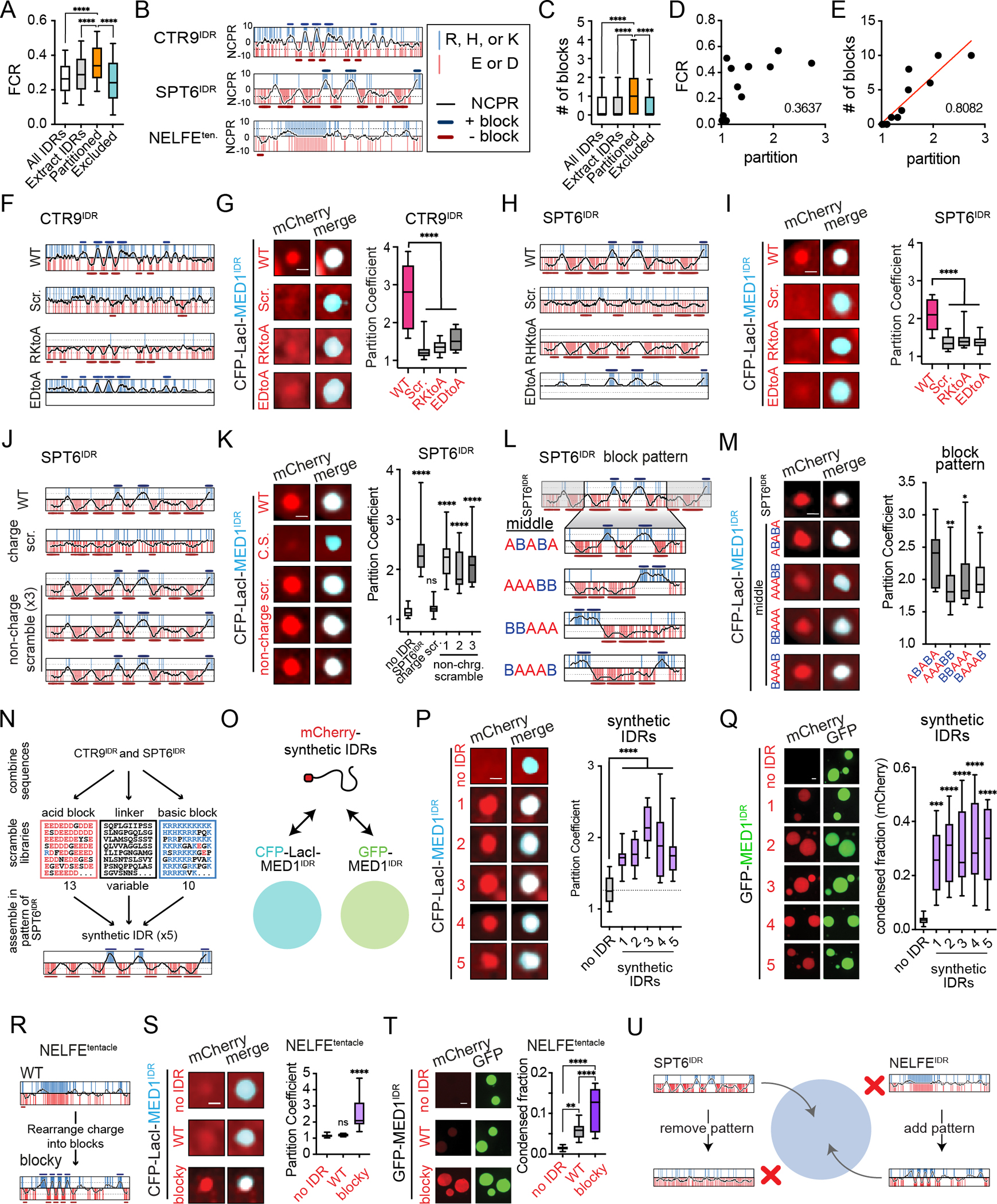
A) Partitioned IDRs have a higher FCR than other IDR classes tested. Boxplots(10–90%). p-values, one-way ANOVA vs partitioned IDRs.
B) NCPR for 10-residue sliding window (black line) across the indicated disordered regions. Vertical red and blue lines represent acidic or basic residues, respectively. Y-axis, charge index. The position of acidic or basic blocks (10-residue window NCPR ≥ abs (5)) is shown as dark red bars or dark blue bars, respectively.
C) Same as 5A, but for number of charged blocks.
D) Correlation analysis of partition coefficient and FCR for all IDRs tested in Lac array cells (Fig 4I, S4O). r2 = goodness-of-fit for a linear regression.
E) Same as 5D but for number of charged blocks. Red line, linear fit.
F) NCPR plots as in 5B are shown for CTR9IDR and the CTR9IDR mutants tested.
G) Left: images of indicated mCherry fusions and CFP-LacI-MED1IDR. Scale, 1μm. Right: Boxplot(10–90%) of partition coefficients for mCherry-CTR9IDR wildtype and mutants shown on left. p-value, one-way ANOVA vs WT.
H) Same as 5F for SPT6IDR.
I) Same as 5G for SPT6IDR.
J) NCPR plots as in 5B for SPT6IDR charge and non-charge scramble mutants.
K) Same as 5G for indicated SPT6IDR mutants.
L) NCPR plots as in 5B for the middle region of SPT6IDR (ABABA pattern) and block pattern rearrangements of this middle region.
M) Same as 5G but for block pattern mutants.
N) Schematic of synthetic IDR design. See methods.
O) Illustration of synthetic IDRs tested for partitioning into CFP-LacI-MED1IDR foci in cells and GFP-MED1IDR droplets in vitro.
P) Same as 5G but for synthetic IDRs.
Q) Left: images and quantification of synthetic IDRs partitioning into GFP-MED1IDR droplets in the presence of RNA. Scale, 2μm. Right: Boxplot(10–90%) of condensed fraction of mCherry signal. p-values, one-way ANOVA vs no IDR control.
R) NCPR plots as in 5B of NELFEtentacle (WT) and NELFE blocky tentacle (blocky).
S) Same as 5G, but with WT and blocky NELFEtentacle.
T) Same as 5Q, but with WT and blocky NELFEtentacle.
U) Illustration summarizing observed sufficiency and necessity of alternating charge block pattern for partitioning into MED1IDR condensates.
See also Figure S5.
