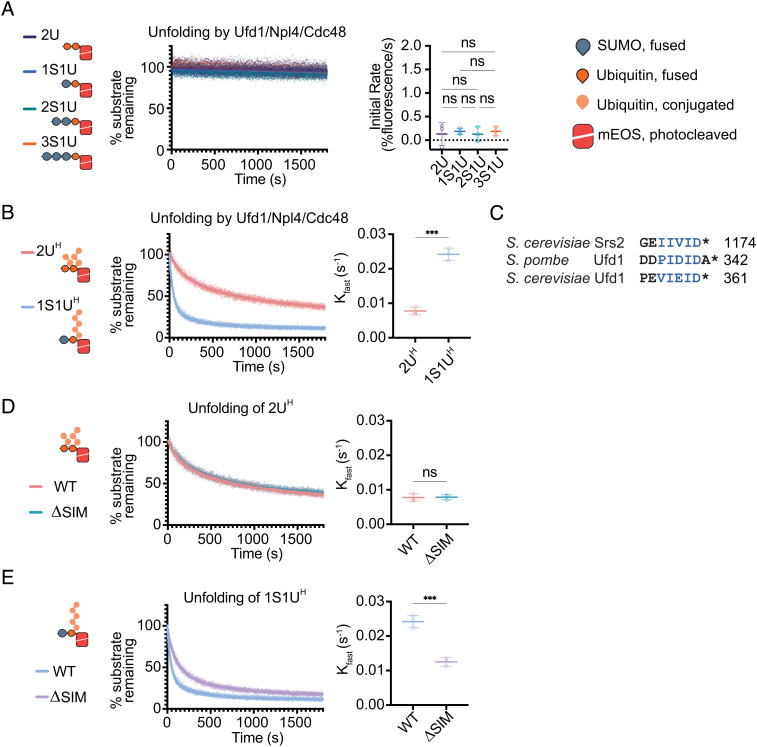
Fig. 2.
Unfolding depends on polyubiquitin, SUMO, and SIM. (A) Non-polyubiquitylated substrates are not readily unfolded. Loss of fluorescence was measured for non-ubiquitylated substrates 2U, 1S1U, 2S1U, and 3S1U by WT Ufd1/Npl4/Cdc48. (B) Unfolding increases in the presence of SUMO. Unfolding of either 2UH or 1S1UH by WT Ufd1/Npl4/Cdc48. (C) Alignment of C-terminal residues of Srs2 from S. cerevisiae, and Ufd1 from S. pombe and S. cerevisiae. Blue lettering indicates SIM. Asterisk (*) denotes the C-terminus of the protein. Unfolding of 2UH (D) or 1S1UH (E) by WT Ufd1/Npl4/Cdc48 (WT) or Ufd1ΔSIM/Npl4/Cdc48 where the SIM was deleted from Ufd1 (ΔSIM). Values were normalized to background fluorescence in the absence of ATP. Plot of three replicates with fit of linear regression (A) or two-phase nonlinear regression (B, D, and E). Initial rate determined using the first 30 s of linear fit or Kfast (s−1) determined using two-phase exponential decay fit. Error bars represent SD. P values calculated by one-way ANOVA with Tukey’s test (A) or unpaired two-tailed t test (B, D, and E); *< 0.05, **P < 0.01, ***P < 0.001, ns (not significant).