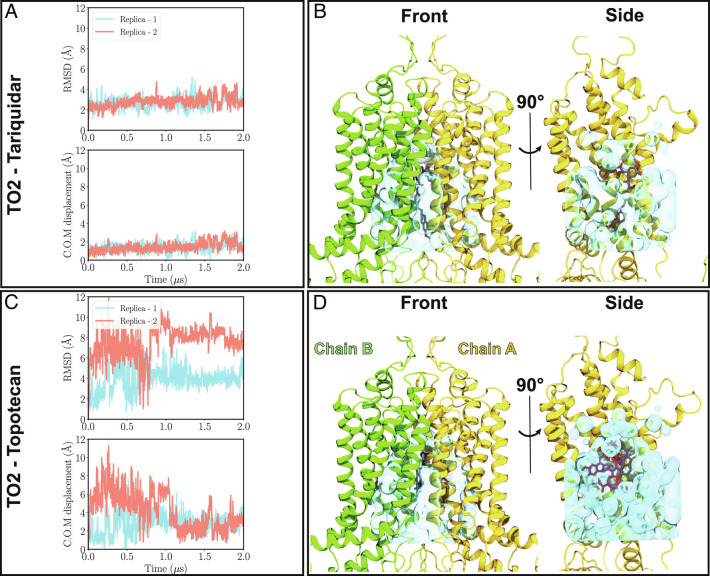
Fig. 2.
Binding stability of tariquidar and topotecan to ABCG2. (A) and (B) Top: Heavy-atom RMSD values for tariquidar and topotecan with respect to their initial positions in the cryo-EM model of the TO2 structure are shown for both simulation replicas (pink and blue traces). While tariquidar is stable in the binding pocket throughout the simulation (RMSD ≈2 Å), topotecan fluctuates extensively, with RMSD values ranging 2 to 12 Å. Bottom, Minimal center of mass distance of tariquidar or topotecan with respect to their two symmetry-related binding poses in the cryo-EM model. (C) and (D) show front and side protein views highlighting the empty volume (shown in cyan) within the binding cavity of ABCG2 for tariquidar and topotecan cryo-EM structures, respectively. Cryo-EM-modeled ligand poses are shown in gray. In the side views, one of the protein chains (chain B) is removed for an unobstructed view of the internal cavity. The larger size of tariquidar leaves less room for fluctuation and therefore a more stable binding mode. In contrast, topotecan’s smaller size allows for more freedom and larger fluctuations of the ligand. Representative topotecan poses arising during the simulations are shown in red in the side view.