Fig. 4.
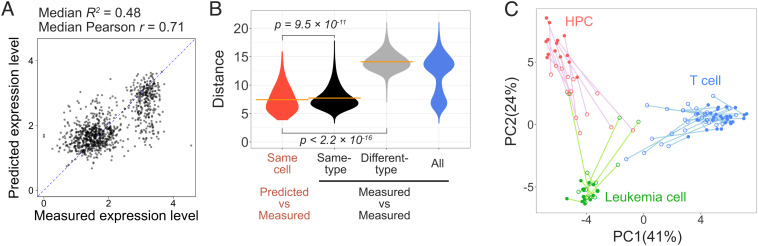
Deep learning–based regression for time-lapse imaged three-type–mixed cells (3mix-ALPS-timelapse). (A) Scatterplots of the measured and predicted RNA expression levels of the cells for the Gm1821 gene (log-transformed; SI Appendix, Text 6), including 16 predictions using different plates. Median R2, median of the out-of-sample R2 value for each prediction (SI Appendix, Fig. S18C); median Pearson’s r, median of the Pearson’s correlation coefficient r for each prediction (SI Appendix, Fig. S18D). Dashed line, diagonal line. (B) Distribution of the Euclidean distances between the predicted and measured expression levels of 300 genes for the same cell and between the measured expression levels of 300 genes of all possible pairs of all cells, same-type cells, and different-type cells. The orange lines indicate the means. The P values were calculated using the Kruskal–Wallis rank sum test. (C) PCA of the cells from 3mix-timelapse-plate5 with predicted (open circle) and measured (closed circle) expression levels of 300 genes (each cell has two dots) (data from the other 15 plates are shown in SI Appendix, Fig. S19). Colors correspond to the three different cell types determined by the measured whole transcriptome. Lines link the predicted and measured expression levels of the same cell.