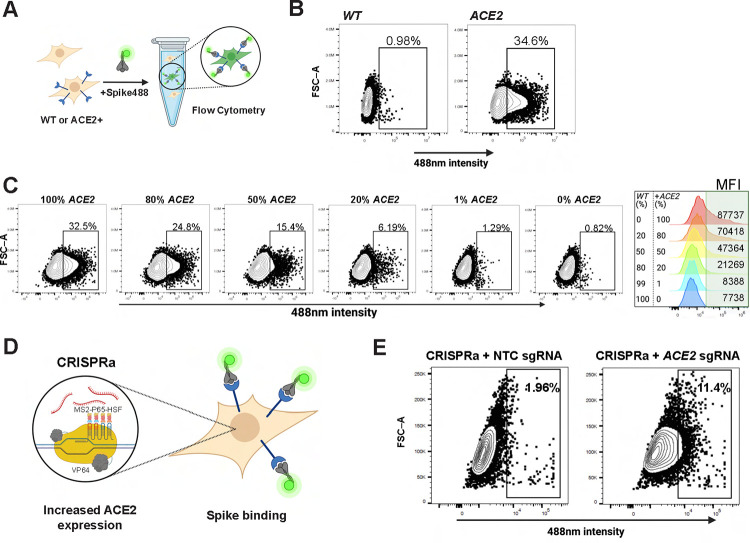
Fig 1. A sensitive FACS-based SARS-CoV-2 spike-binding assay amenable to high-throughput screening.
(A) Schematic of SARS-CoV-2 spike-binding assay. HEK293T cells with stable integration of ACE2 cDNA for overexpression (HEK293T-ACE2) are incubated with Alexa Fluor 488-conjugated SARS-CoV-2 spike protein (Spike488). Spike488-binding cells are then detected by flow cytometry. (B) Representative flow cytometry plots for WT HEK293T and HEK293T-ACE2 incubated with Spike488 (N = 3). See also S1B Fig for gating strategy. (C) Titration of HEK293T-ACE2 (ACE2) cells with WT HEK293T cells. Approximately 1% HEK293T-ACE2 cells showed a difference to baseline non-specific binding. Histogram summary showing MFI of flowed cells. (D) Schematic of CRISPRa system used. (E) Representative plot of flow cytometry for a clonal HEK293T-CRISPRa cell line transduced with NTC sgRNA or ACE2 sgRNA (expression confirmation via RT-qPCR in S1A Fig). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7416876. ACE2, angiotensin-converting enzyme 2; CRISPRa, CRISPR activation; MFI, mean fluorescence intensity; NTC, non-targeting control; SARS‑CoV‑2, Severe Acute Respiratory Syndrome Coronavirus 2; sgRNA, single-guide RNA.