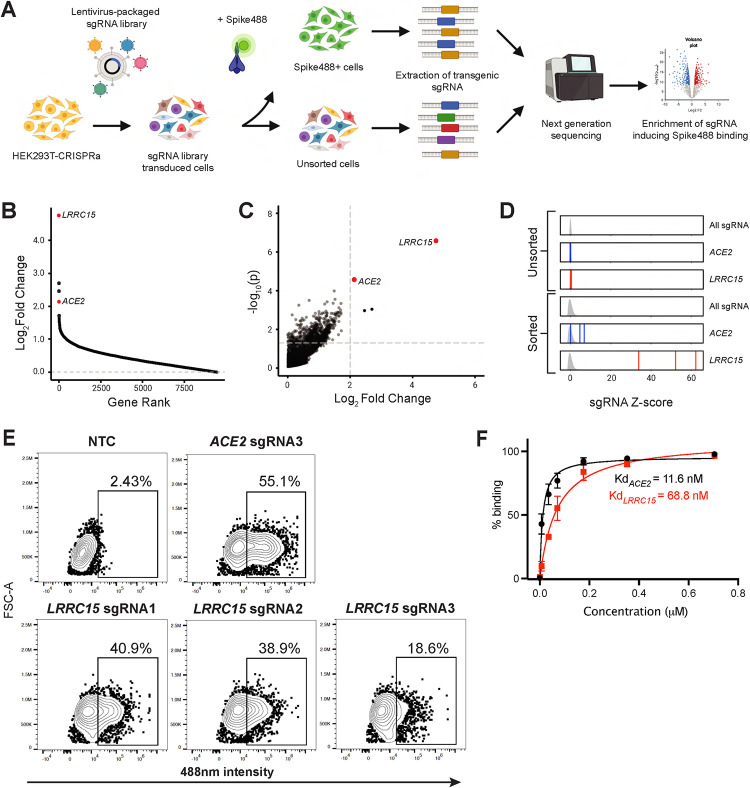
Fig 2. Whole-genome CRISPRa screening identified LRRC15 as a novel SARS-CoV-2 spike-binding protein.
(A) Schematic of CRISPRa screen used to identify regulators of SARS-CoV-2 spike binding. (B) Ranking of all genes in screen 1 by log2 fold change (LFC) calculated using MAGeCK and plotted using MAGeCKFlute. See also S1 Table. (C) Gene enrichment analysis of screen 1 performed using MAGeCK. Horizontal dotted line indicates p-value = 0.05. Vertical dotted line indicates LFCs of 2. P-values and LFCs for all genes are reported in S1 Table. (D) sgRNA Z-scores for screen 1 unsorted and sorted samples. Density curve for all sgRNA Z-scores in sample (i.e., sorted or unsorted) is shown in gray. Z-scores for each guide are indicated by vertical lines (blue ACE2, red LRRC15). (E) Flow cytometry analysis of HEK293T-CRISPRa cells transduced with 3 independent LRRC15 sgRNAs. HEK293T-CRISPRa transduced with ACE2 sgRNA3 were used as a positive control and NTC sgRNA-transduced HEK293T-CRISPRa cells were used as a negative control (N = 3). (F) Quantification of Spike647 binding in ACE2 sgRNA3 and LRRC15 sgRNA1 cells via flow cytometry. Dissociation constant (Kd) was calculated by fitting with nonlinear regression (1 site—specific binding). N = 3, error bars represent SD. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7416876. ACE2, angiotensin-converting enzyme 2; CRISPRa, CRISPR activation; LFC, log2 fold change; LRRC15, leucine-rich repeat-containing protein 15; NTC, non-targeting control; SARS‑CoV‑2, Severe Acute Respiratory Syndrome Coronavirus 2; sgRNA, single-guide RNA.