Figure 5: Diversity of HDV antigen-like proteins in known ribozyviruses and ribozy-like viruses identified in metatranscriptomes.
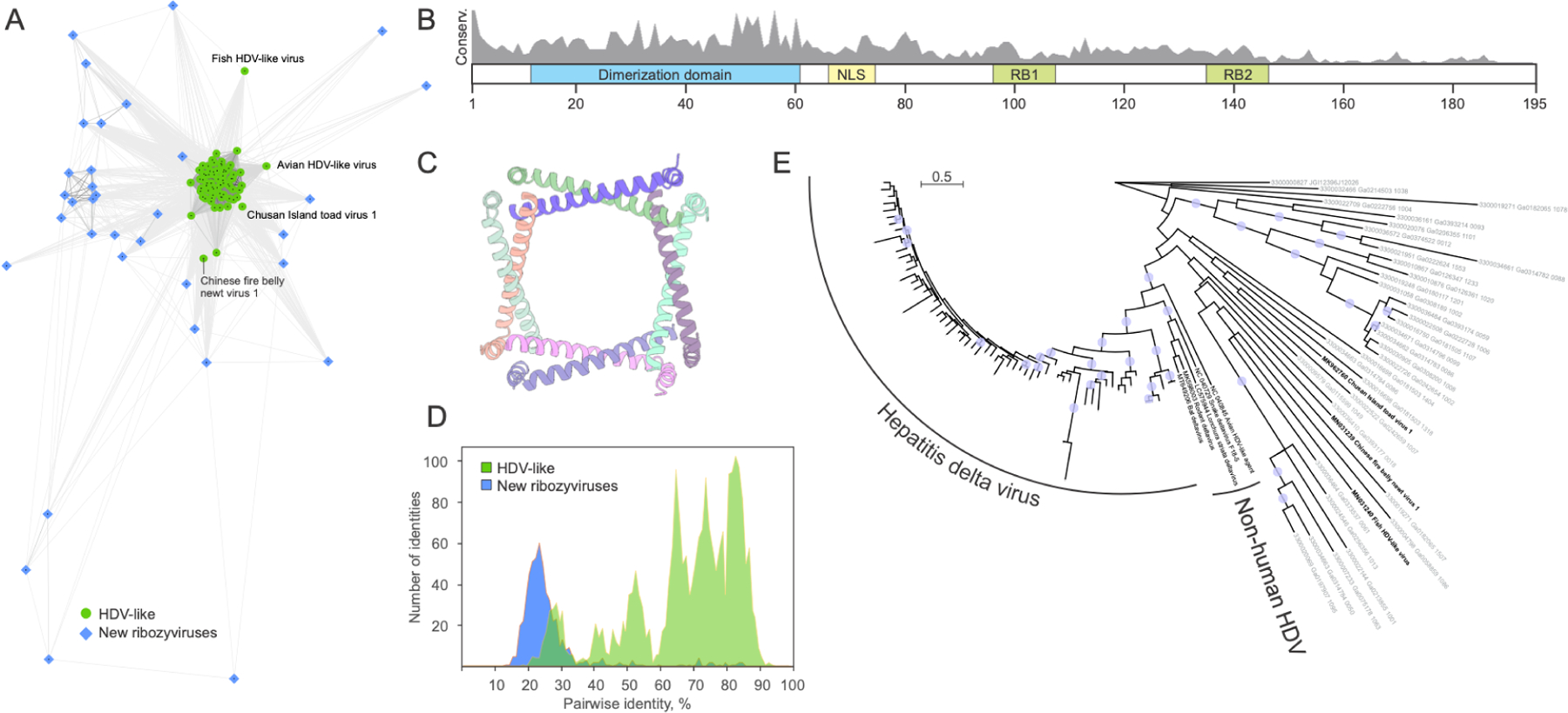
A. Clustering of the HDV antigen (Ag)-like protein homologs based on their sequence similarity. Lines connect nodes (sequences) with P-value < 1e-05. Reference HDVAg-like sequences from GenBank are shown as green circles, whereas those detected in metatranscriptomic datasets as blue diamonds. Some of the divergent reference sequences are labeled. B. Schematic representation of the HDVAg with functionally important regions indicated with colored boxes. RB1 and RB2, RNA-binding sites 1 and 2, respectively; NLS, nuclear localization signal. Gray histogram shows the sequence conservation (percent identity) of HDVAg-like sequences from metatranscriptomic datasets. C. Octameric structure of the conserved dimerization domain of HDVAg. PDB ID: 1A9262. Each protein molecule is shown with a different color. D. Comparison of the sequence conservation among reference HDVAg from GenBank (green) and those from metatranscriptomic datasets (blue). E. Maximum likelihood phylogeny of HDVAg-like sequences. The tree was constructed with IQ-TREE63. Circles at the nodes represent SH-aLRT support higher than 90%. The scale bar represents the number of substitutions per site.
