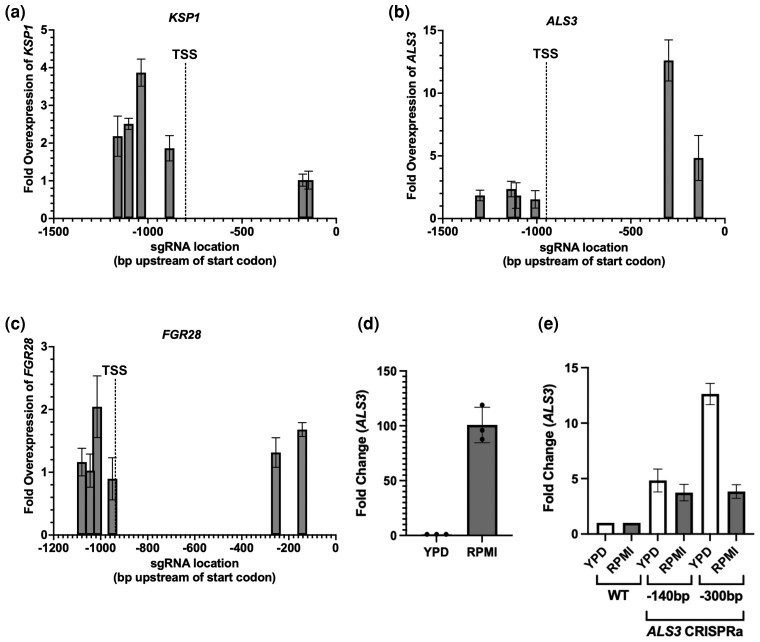
Fig. 2.
Determining optimal guide-design principles for CRISPRa overexpression in Candida albicans. CRISPRa strains were created using 6 different sgRNA molecules designed to target different positions in the promoter region of each of a) KSP1, b) ALS3, and c) FGR28 upstream of either the start codon or the predicted dominant TSS. Fold change in the expression of the target gene relative to the housekeeping gene ACT1 in the experimental strains and the nontargeting CRISPRa control strain was measured with RT-qPCR and compared. Data points represent the mean fold difference and standard deviation in expression of the respective target gene in the CRISPRa strains compared with the nontargeting CRISPRa control (n = 3). d) Fold change in the basal expression level of ALS3 relative to the housekeeping gene ACT1 in the nontargeting CRISPRa control strain grown in either standard laboratory conditions (YPD at 30°C) or conditions known to favor a high level of ALS3 expression (RPMI at 37°C) was measured with RT-qPCR. Data represent the mean fold difference and standard deviation in expression of ALS3 in the nontargeting CRISPRa control strain in both conditions normalized to growth in YPD at 30°C (n = 3). e) Fold change in the expression of ALS3 relative to the housekeeping gene ACT1 in two CRISPRa strains targeting ALS3 for overexpression as well as the nontargeting CRISPRa control strain grown in either YPD at 30°C or RPMI at 37°C was measured with RT-qPCR. Data represent the mean fold difference and standard deviation in expression of ALS3 in all strains in both conditions normalized to the nontargeting CRISPRa control strain (WT; n = 3).