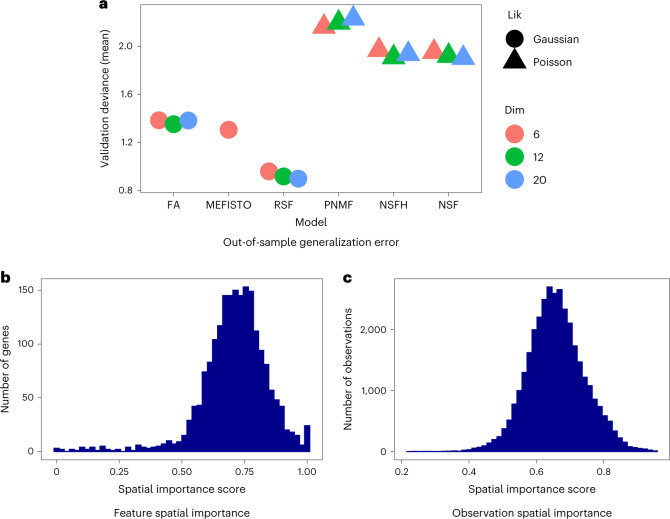
Fig. 2. Benchmarking spatial and nonspatial factor models on Slide-seqV2 mouse hippocampus spatial gene expression data.
a, Poisson deviance on held-out validation data. Lower deviance indicates better generalization accuracy. All spatial models used 2,000 IPs. MEFISTO could not be fit with more than six components due to out of memory errors. Lik represents likelihood, and Dim represents the number of latent dimensions (components). b, Each feature (gene) was assigned a spatial importance score derived from NSFH fit with 20 components (ten spatial and ten nonspatial). A score of 1 indicates spatial components explain all the variation. c, As b but with observations instead of features.