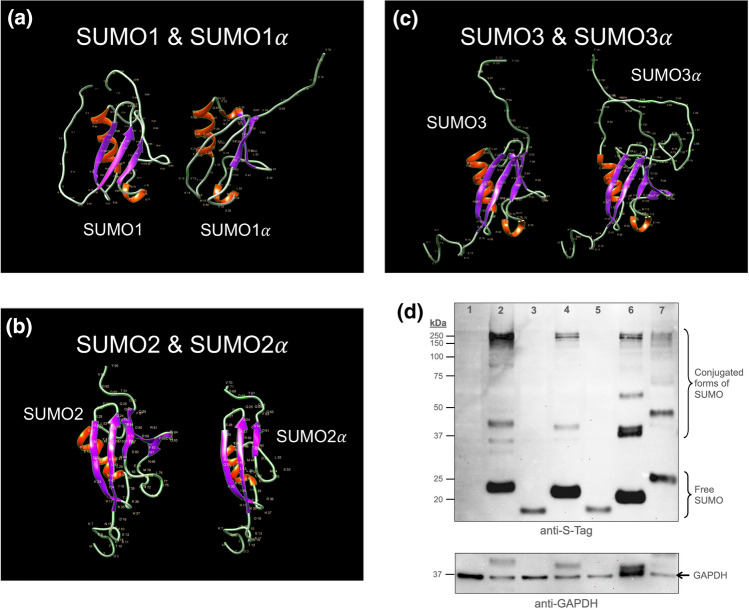
Figure 8.
The exon-deficient SUMO alphas (SUMO1α and SUMO2α) are non-conjugatable. (a–c). Crystal structure of the SUMO alphas as compared to those of their prototypical isoform as predicted by RaptorX. The images shown were considered to best emphasize the differences in predicted crystal structures. Notice how the additional sequence present in SUMO3α is predicted to form an unstructured region that remains outside the central globular core of SUMO3α, therefore imposing minimal disruptions in the globular core; in sharp contrast, the specific amino acid sequence deletions present in SUMO1α and SUMO2α are predicted to trigger substantial changes in their globular core as compared with their prototypical isoforms. Virtually identical tertiary structures were predicted by Alpha Fold analyses. (d) Immunoblot analyses showing the patterns of conjugation associated to each SUMO isoform. HEK293A cells were transfected with expression constructs coding for the indicated His-S-tagged SUMO protein. At 24 h post-transfection, the cells were lysed and the resulting extracts analyzed by SDS-PAGE and immunoblotting. Upper panel: immunoblot performed with antibodies directed against the S-tag (located near the N-terminal end of the proteins). Lower panel: subsequent immunoblot using anti-GAPDH antibodies as loading control for the different samples. The localization of free- and conjugated-SUMO forms is indicated. High molecular weight signals are easily visible for SUMO3α, indicating its ability to become conjugated to other proteins. In contrast, no high molecular weight forms were observed for SUMO1α and SUMO2α. 1, Mock transfected cells; 2 and 3, cells over-expressing SUMO1 and SUMO1α, respectively; 4 and 5, cells over-expressing SUMO2 and SUMO2α, respectively; 6 and 7, cells over-expressing SUMO3 and SUMO3α, respectively. All over-expressed SUMO proteins contained tandem His- and S-tags at their N-terminus.