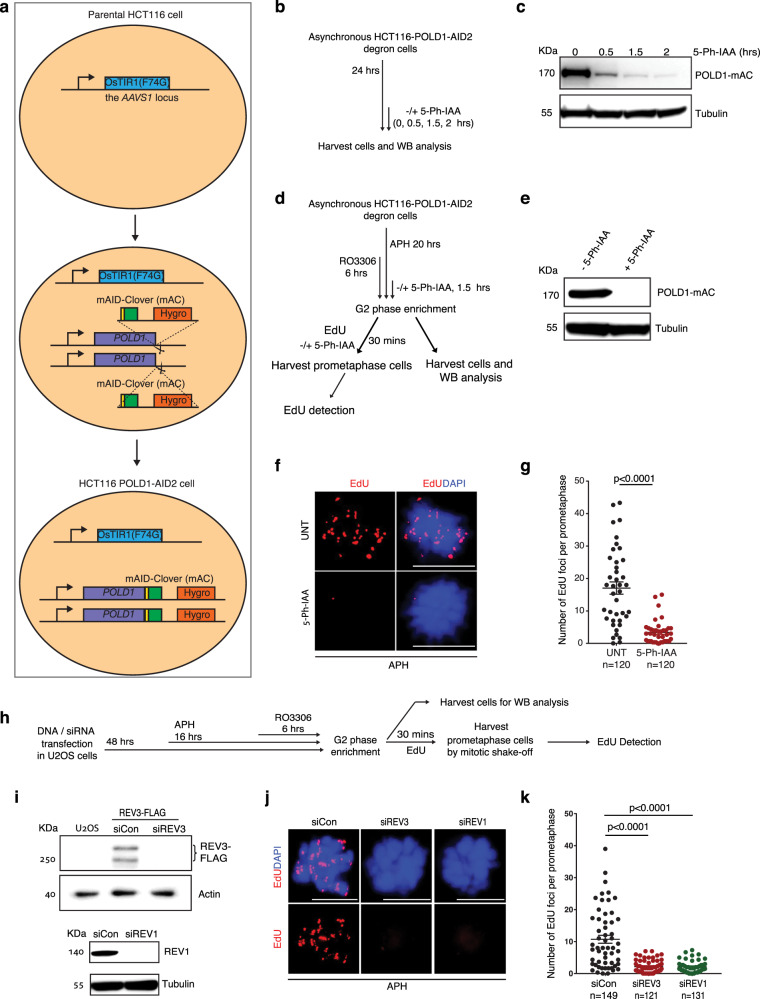
Fig. 1. POLD1, REV3 and REV1 are essential for MiDAS.
a Workflow for establishing an HCT116-POLD1-AID2 cell line expressing only POLD1 that is tagged with OsTIR1(F74G) (mAC). b Experimental workflow for assessing POLD1 protein degradation following different periods of incubation with 5-Ph-IAA. c Western blot (WB) analysis of POLD1-mAc in asynchronous cells treated as shown in panel b. β-tubulin was a loading control. d Experimental workflow for analysis of MiDAS in prometaphase HCT116-POLD1-AID2 cells following APH treatment, enrichment of G2 cells by RO3306, and POLD1-mAC degradation. POLD1-mAC depletion was induced by treating cells with 5-Ph-IAA for 2 h. e WB analysis of POLD1-mAC (detected with a POLD1 antibody) following 2 h 5-Ph-IAA treatment. β-tubulin was used as a loading control. Representative images (f) and quantification (g) of MiDAS foci (labeled with EdU; red) in prometaphase cells treated as shown in panel d. h Experimental workflow for U2OS cell synchronization and analysis of MiDAS in prometaphase cells following REV3 or REV1 depletion. In b and h, before being incubated with EdU for 30 min (with or without 5-Ph-IAA), cells were rinsed three times with pre-warmed, drug-free culture medium within 5 min. i Top, WB analysis of REV3FLAG following transfection of a REV3FLAG expressing plasmid together with control or REV3 siRNAs. Flag antibody was used to detect flag-REV3, since a reliable REV3 antibody is not available. Actin was used as a loading control. Protein sample from untransfected U2OS cells was used as a negative control. Lower, WB analysis of REV1 after transfecting cells with control or REV1 siRNAs. β-tubulin was used as a loading control. Representative images (j) and quantification (k) of MiDAS foci (labeled with EdU; red) in prometaphase cells treated as shown in panel h. DNA was stained with DAPI (blue). Each data point in charts of g and k is means of three independent experiments and plotted with Prism (n = number of cells analyzed in each condition in three independent experiments). Error bars represent SEM. P values were calculated using a two-tailed non-parametric Mann–Whitney test. Scale bars, 10 μm. Hr hour, min minute.