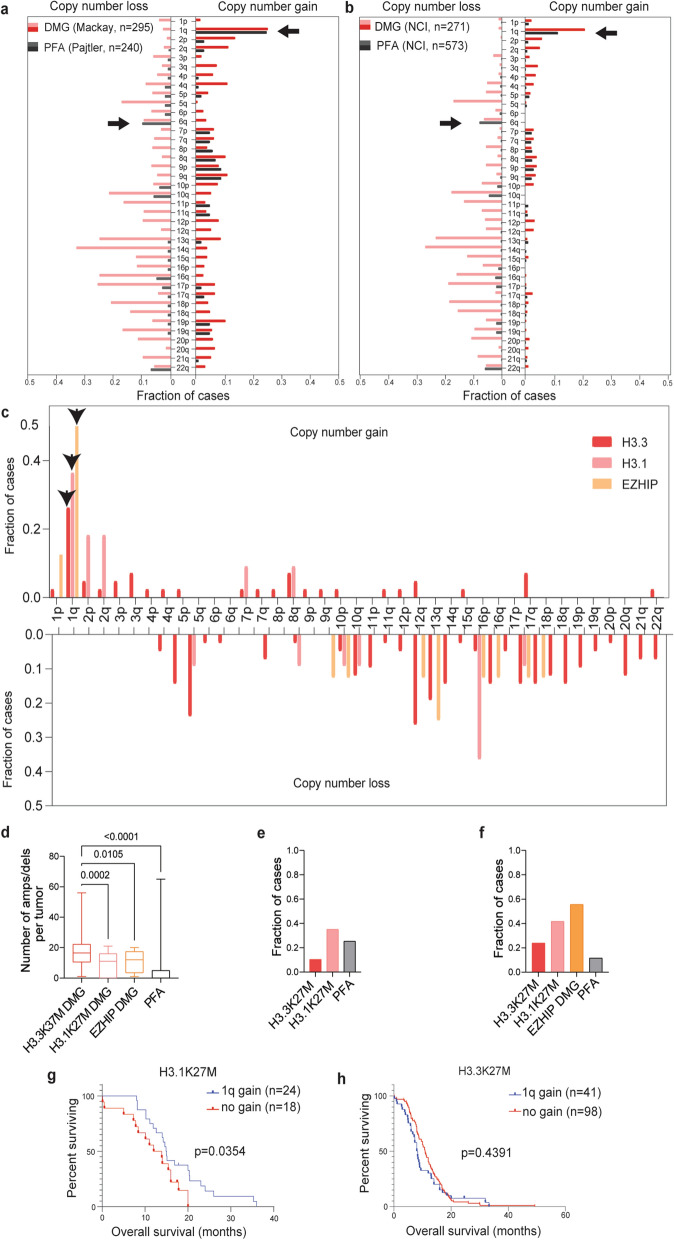
Fig. 1.
H3.1K27M and EZHIP-DMGs share key copy-number alterations with PFAs. a Copy-number profiles (loss left panel, gain right panel) of H3K27M DMGs (Mackay et al. 2017, n = 295) [36] and PFAs (Pajtler et al. 2015, n = 240) [37] from published DNA methylation cohorts. Arrows indicate shared recurrent alterations (more than 5% of tumors in all cohorts) in a and b with known survival associations in PFAs. X-axis = fraction of cases and Y-axis = chromosomes. b Copy-number profiles (loss left panel, gain right panel) of H3K27-altered DMGs (n = 271) and PFAs (n = 573) from the National Cancer Institute (NCI). Arm-level copy-number gain/loss calls were made using GISTIC score cutoffs of ± 0.3. c Frequency of chromosome arm-level copy number alterations (gain top panel, loss bottom panel) for H3K27-altered DMGs from b segregated by molecular alterations: H3.3 mutant (n = 42), H3.1 mutant (n = 11) and H3-wildtype (H3-WT), EZHIP-expressing (n = 8). X-axis = chromosomes and Y-axis = fraction of cases. d Quantification of the frequency of amplifications (amps) and deletions (dels) (Y-axis) in DMGs segregated by molecular alterations (X-axis) and in PFAs from the NCI cohort. Data analyzed by one-way ANOVA with 95% confidence intervals. H3.3 mutant (n = 48), H3.1 mutant (n = 12), and EZHIP (n = 8). e Frequency of 1q gain (Y-axis) for subtypes of H3K27M DMG and PFAs from published datasets depicted in a. H3.3 mutant (n = 245), H3.1 mutant (n = 49), and PFA (n = 240). DMG sample pHGG_META_0223 was excluded from as it is H3.2-mutant. f Frequency of 1q gain (Y-axis) for subtypes of H3K27-altered DMG and PFA ependymoma from the NCI cohort depicted in 1b. H3.3 mutant (n = 42), H3.1 mutant (n = 11), and EZHIP (n = 8). g Overall survival (months) analysis of 1q gain in H3.1K27M DMGs with (n = 24) or without (n = 18) 1q gain. h Overall survival (months) analysis of 1q gain in H3.3K27M DMGs with (n = 41) or without (n = 98) 1q gain. Data in g–h analyzed using Kaplan–Meier with Log-Rank test with 95% confidence intervals