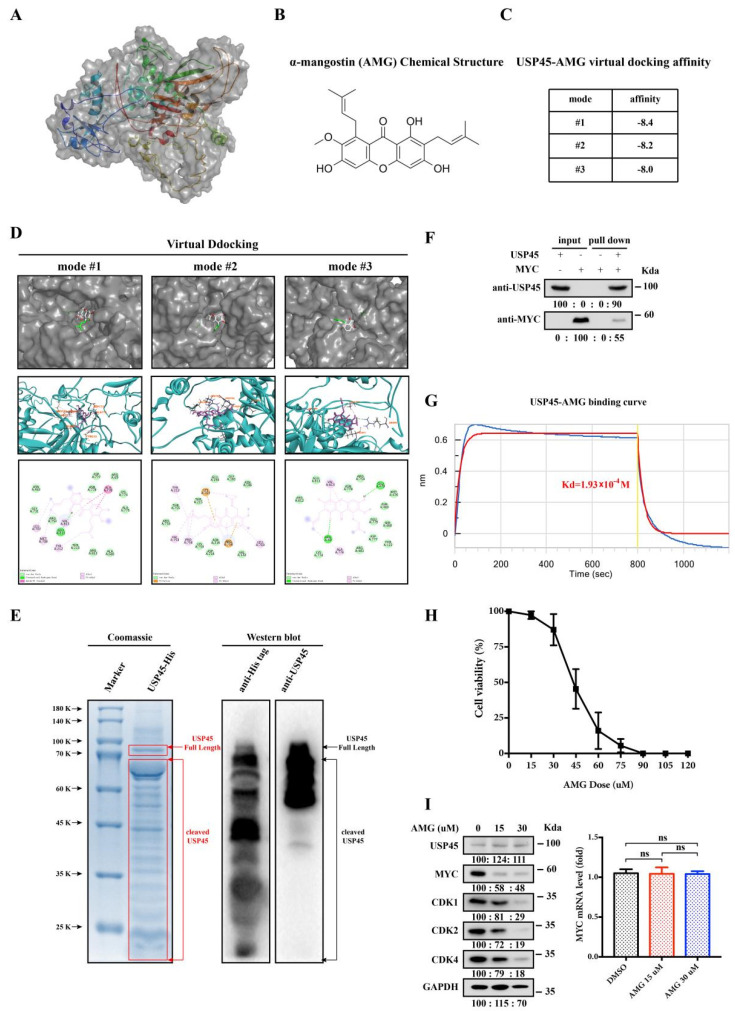
Figure 5.
Alpha-mangostin (AMG) may inhibit the deubiquitination-active sites of USP45. (A) The structure of USP45 protein was simulated with I-TASSER program. (B) AMG chemical structure. (C) Top three scores of USP45-AMG virtual docking affinities. (D) Interactions between USP45 and AMG: the partial USP45-AMG docking structure (top), 3D diagram (middle) and 2D diagram (bottom) of AMG interaction with the amino acids of USP45. (E) USP45 purified from E. coli was analyzed by Coomassie and Western Bolt. (F) USP45 purified from E. coli and MYC purified from HEK293T cells were incubated together, followed by pulling down. (G) The USP45-AMG affinity was determined using BLI-OCTET K2. (H) SiHa cells were treated with different AMG dosages for 48 h, and the cell viability was determined by MTT assay. (I) SiHa cells were treated with different dosages of AMG for 48 h and subjected to Western Blot and qPCR analyses, DMSO as the negative control. Experiments were conducted at least two times in duplicates.