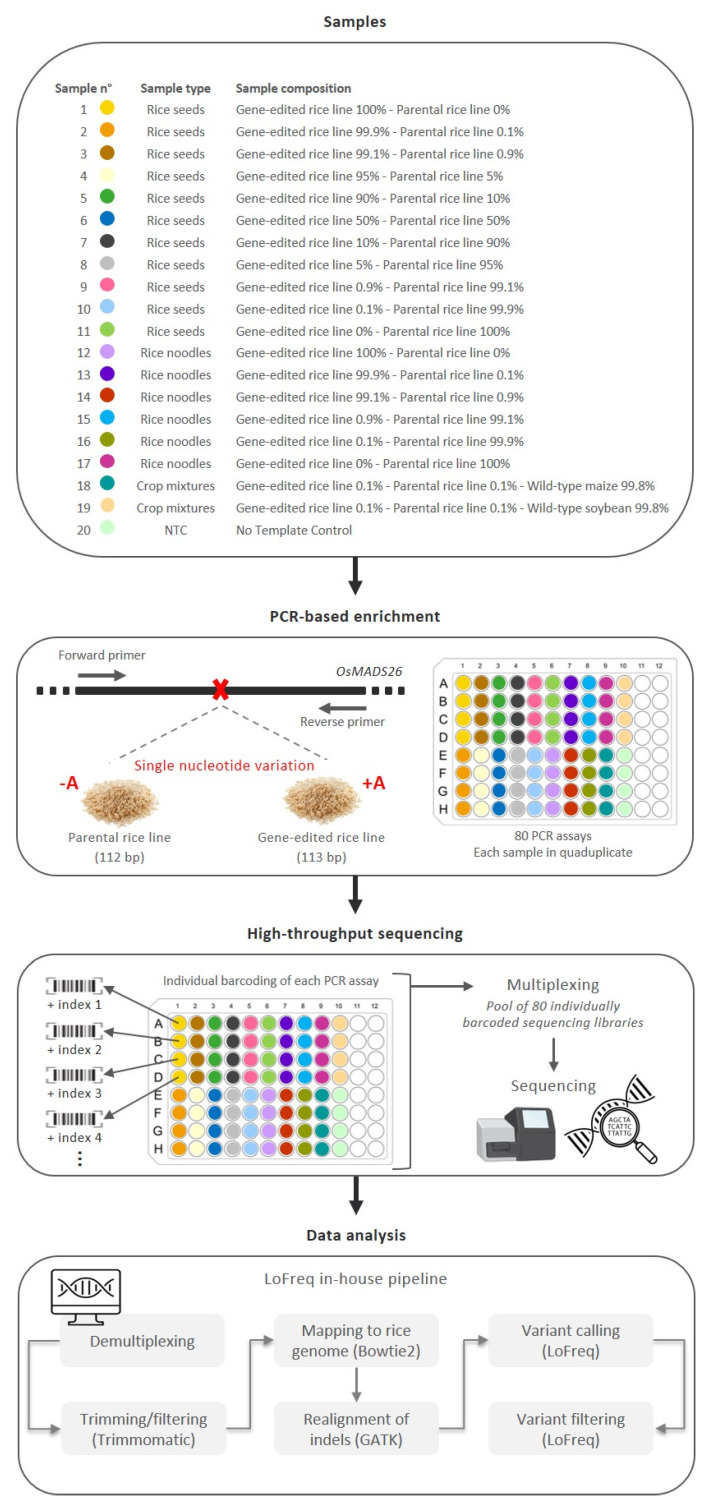
Figure 1.
Schematic workflow of the targeted high-throughput sequencing approach performed in this study. A total of 20 different samples were analyzed using the proposed targeted high-throughput sequencing approach, including 19 samples containing various percentages of the GE rice line as well as 1 NTC. The proposed targeted high-throughput sequencing approach is composed of three successive steps. First, a PCR-based enrichment step is applied to the whole DNA extracted from a given sample. The used primer pair was previously designed to amplify the OsMADS26 region carrying an SNV of interest (insertion or deletion of a single adenosine), resulting in a PCR amplicon of 113 bp for the GE rice line and a PCR amplicon of 112 bp for its parental rice line [16]. For each of these 20 samples, the PCR assay was performed in quadruplicate, resulting in a total of 80 final PCR products. Second, each PCR product is individually coupled to a unique barcode and then pooled for high-throughput sequencing. Third, raw sequencing reads are analyzed using an in-house pipeline based on LoFreq, developed to identify SNVs as well as to estimate their frequency.