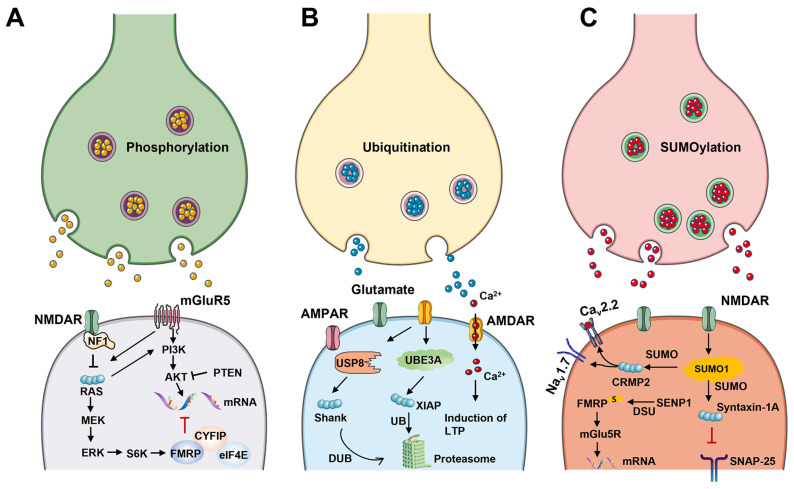
Figure 4.
A network of epigenetic post-translational modifications associated with ASD pathophysiology. (A) Ras inactivation is generally caused by neurofibrillar proteins encoded by NF1. Neurofibrillary protein mutations in neurofibromatosis 1 cause excessive activation of the Ras/ERK and PI3K signaling pathways, affecting and inhibiting translation of proteins encoded by genes associated with neurodegenerative diseases in autism. (B) USP8 is required for synaptic function by stabilizing Shank family proteins to prevent degradation. UBE3A regulates synaptic activity by targeting XIAP ubiquitination. (C) FMRP activity-dependent SUMOylation is a critical step in the separation of FMRP from dendritic mRNA particles, which regulates spine elimination and maturation. CRMP2 is a SUMO substrate that reduces Ca2+ entry via the presynaptic voltage-gated Ca2+ channel CaV2 in a dynamic manner. 2. CRMP2 SUMOylation is also thought to regulate membrane expression in the sodium channel NaV1.7. Syntaxin-1A SUMOylation is induced by NMDAR activation, resulting in decreased binding to SNAP-25 and serving as a key presynaptic modulator of vesicular endocytosis.