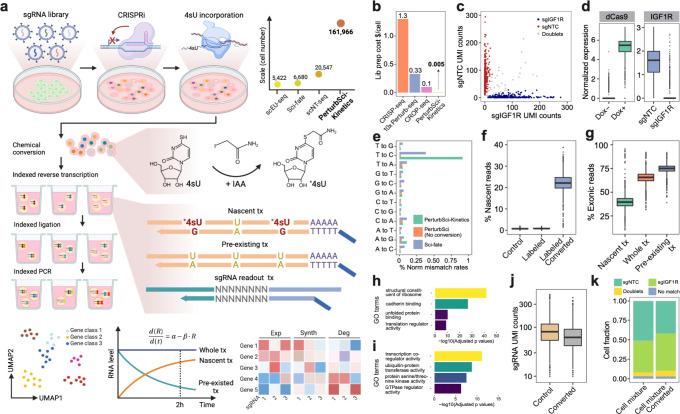
Fig. 1. PerturbSci-Kinetics enables joint profiling of transcriptome dynamics and high-throughput gene perturbations by pooled CRISPR screens.
a. Scheme of the experimental and computational strategy for PerturbSci-Kinetics. The dot plot on the upper right shows the number of cells profiled in this study for comparison with the published single-cell metabolic profiling datasets14,15,24. Scale, the highest number of cells profiled in a single experiment of each technique. IAA, iodoacetamide. *4sU, chemically modified 4sU. R, steady-state RNA level. α, RNA synthesis rate. β, RNA degradation rate. Exp, steady-state expression. Synth, synthesis rate. Deg, degradation rate. b. Bar plot showing the estimated library preparation cost for PeturbSci-Kinetics and other published techniques25,26 for single-cell transcriptome analysis coupled with CRISPR screens. c. Scatter plot showing the number of unique sgRNA transcripts detected per cell in the PerturbSci experiment for profiling cells transduced with sgNTC or sgIGF1R. d. The left box plot shows the normalized expression of dCas9-KRAB-MeCP2 in untreated or Dox-induced HEK293-idCas9 cells. The right box plot shows the normalized expression of IGF1R in Dox-induced HEK293-idCas9 cells transduced with sgNTC or sgIGF1R. Gene counts of each single cell were normalized by the total gene count, multiplied by 1e4 and then log-transformed. e. Bar plot showing the normalized percentage of all possible single base mismatches in reads from sci-fate (blue), and PerturbSci-Kinetics on chemically converted (green) or unconverted cells (orange). Normalized mismatch rates, the percentage of each type of mismatch in all sequencing bases. f. Box plot showing the fraction of recovered nascent reads in single-cell transcriptomes across conditions: no 4sU labeling + no chemical conversion, 4sU labeling + no chemical conversion, and 4sU labeling + chemical conversion. g. Box plot showing the ratio of reads mapped to exonic regions of the genome in nascent reads, pre-existing reads, and reads of the whole transcriptomes across single cells. h-i. Bar plots showing the significantly enriched Gene Ontology (GO) terms in the list of genes with low (h) or high (i) nascent reads ratio (Methods). j. Box plot showing the number of unique sgRNA transcripts detected per cell in cells with or without the chemical conversion. k. We performed PerturbSci-Kinetics experiment using converted/unconverted HEK293-idCas9 cells transduced with sgNTC/sgIGF1R. Stacked bar plot showing the fraction of converted/unconverted cells identified as sgNTC/sgIGF1R singlets, doublets, and cells with no sgRNA detected.