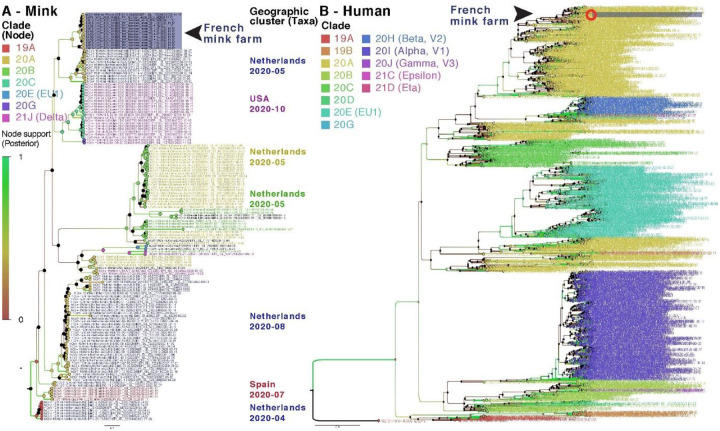
Figure 2.
Bayesian phylogenies on a mix of representative set of SARS-CoV-2 sequences available in GISAID. SARS-CoV-2 sequences detected in the mink farm in France are highlighted in purple. A. Bayesian phylogeny of all SARS-CoV-2 genomes from minks over the study period. Node and taxa labels are colored according to nextstrain (https://nextstrain.org) clade classification and main geographic clusters, respectively. B. Bayesian phylogeny of human derived SARS-CoV-2 genomes collected in France and mink derived SARS-CoV-2 genomes described in this study. A diversity optimized dataset was obtained by collecting and filtering complete, high quality genomes, of SARS-CoV-2 detected in France (of Human and mink origin). Taxa are colored according to nextstrain clades classification.