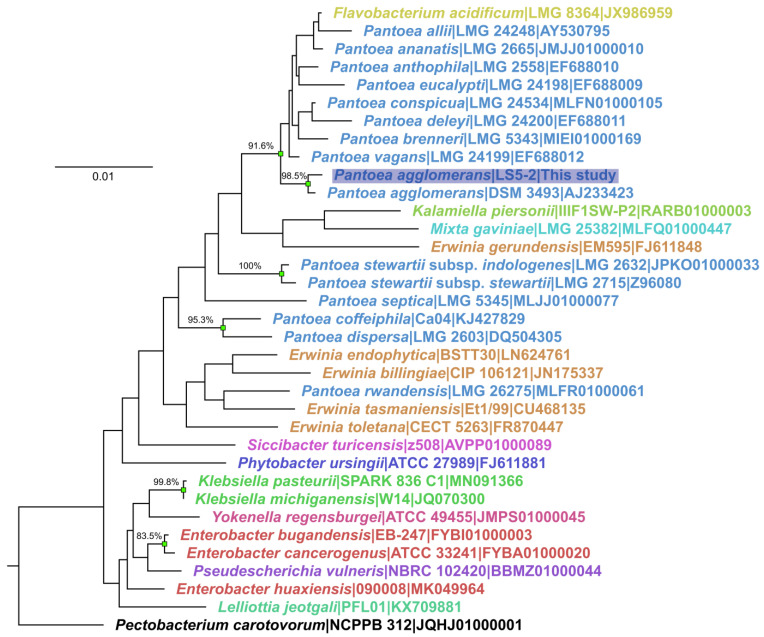
Figure 1.
Neighbor-joining tree of 16S rRNA gene sequences of bacterial isolate LS5-2 and closely related bacterial species. The analysis involved 35 nucleotide sequences (16s rRNA gene sequences of P. agglomerans isolate LS5-2-host strain of the phages described within this study, 33 other closely related bacterial species, and Pectobacterium carotovorum as an outgroup for rooting). Tip labels correspond to the taxa and are in the format of “Species|Strain|Accession”; tip label colors correspond to different bacterial genera. The tip label of the sequence corresponding to the isolate LS5-2 is additionally highlighted in blue. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (out of 1000 replicates) is indicated for branches having bootstrap support higher than or equal to 80%; such branches also have their distal nodes indicated by green rectangles. The evolutionary distances are in the number of base differences per site and the tree is drawn to scale. All positions with less than 90% site coverage were eliminated. That is, fewer than 10% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 1346 positions in the final dataset.