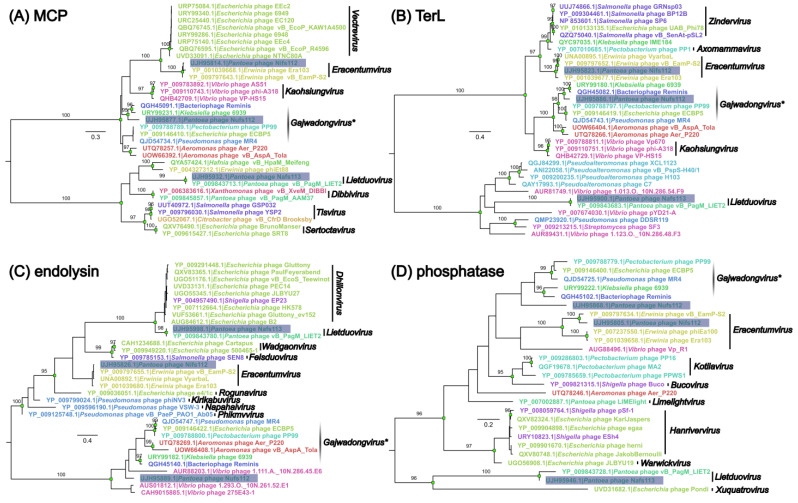
Figure 3.
Midpoint-rooted maximum likelihood trees of the selected studied phage protein amino acid sequences and similar sequences found in the proteomes of other cultured phages. (A) Major capsid protein (MCP), (B) terminase large subunit (TerL), (C) endolysin, (D) phosphatase dataset; multiple sequence alignment and tree details can be found in Supplementary Table S4. The trees are drawn to their respective scales, and branch lengths correspond to the number of amino acid substitutions per site. Branches with UFBoot support of ≥95% (out of 1000 replicates) have their distal nodes indicated as green rectangles, and the corresponding UFBoot value is indicated above the branches. In all of the trees, tips are labeled as “Respective protein accession|Originating phage”, and tip labels are colored based on the respective phage host genus; labels of the leaves containing the amino acid sequences of the respective studied phage proteins are highlighted in blue. Labeled black bars after the tip labels show clade genus level annotations retrieved from the respective phage complete-genome metadata at the time of writing (if indicated). Cloudy black bar labeled “Gajwadongvirus*” indicates dubious genus-level designation for some of the phages and contains some phages without genus-level associated taxonomy information.