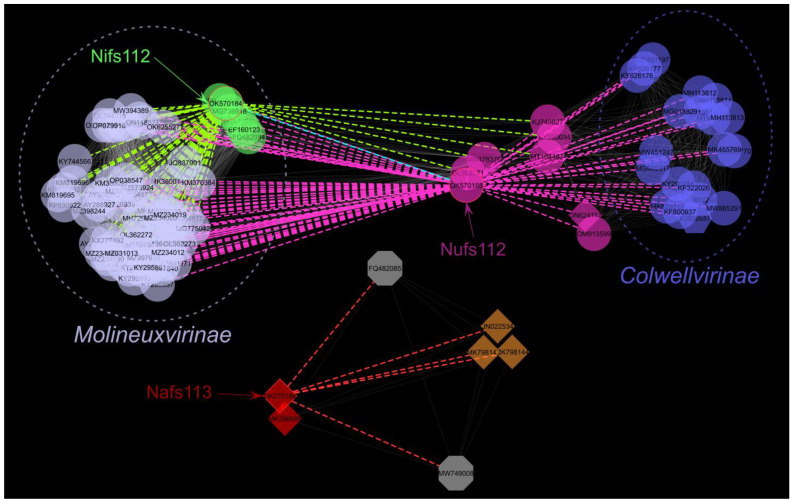
Figure 4.
Sub-network containing first neighbors of the studied phages under edge-weighted spring-embedded layout (n = 103). Nodes are colored according to the viral cluster as identified by vConTACT2 for a network generated from 18,553 complete phage genomes: VC_436_0 (n = 62)—light blue; VC_437_0 (n = 6)—green; VC_961_0 (n = 20)—dark blue; VC_62_0 (n = 3)—brown; VC_794_0 (n = 2)—red; outliers (n = 2)—grey; phages that were determined to be classified as an overlap between two viral clusters (VC_439/VC_962; n = 8) that were not present in the sub-network—magenta. Different node shapes correspond to different viral families: circle—Autographiviridae; octagon—Myoviridae (recently abolished); hexagon—Drexlerviridae; diamond—unclassified at the family level. Dashed ellipses enclose recognized and tentative Molineuxvirinae (light blue) or Colwellvirinae (dark blue) phage subfamily representatives. Dashed colored edges represent links to the studied phages: green—Nifs112; magenta—Nufs112; red—Nafs113. Mutual link between Nifs112 and Nufs112 is represented by cyan dashed line.