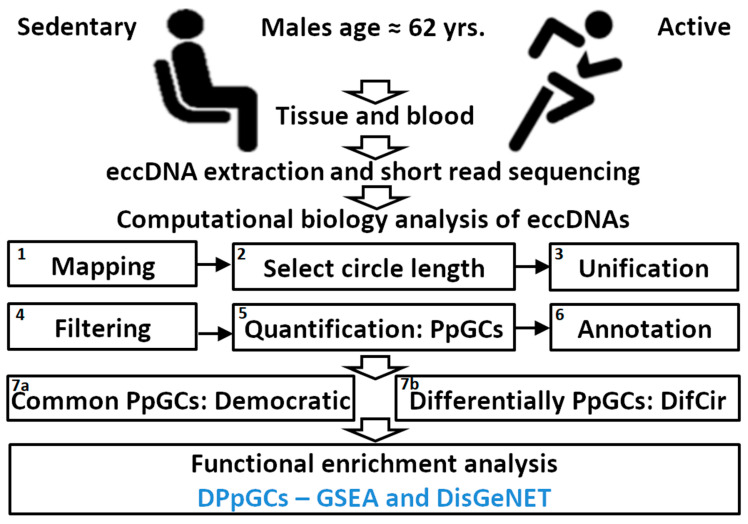
Figure 1.
Experimental setup and computational analysis workflow. Isolation and purification of circular DNA from skeletal muscle (SkM) tissue (T) and blood (B) of sedentary (S) and active (A) individuals and subsequent assembly, annotation, quantification of eccDNA species, quantification of produced per gene circles (PpGCs), calculation of differentially PpGCs (DPpGCs), and identification of common PpGCs (CPpGCs) in the TS and TA groups using a democratic vote method. Functional enrichment analysis of the DPpGCs performed with GSEA and DisGeNET.