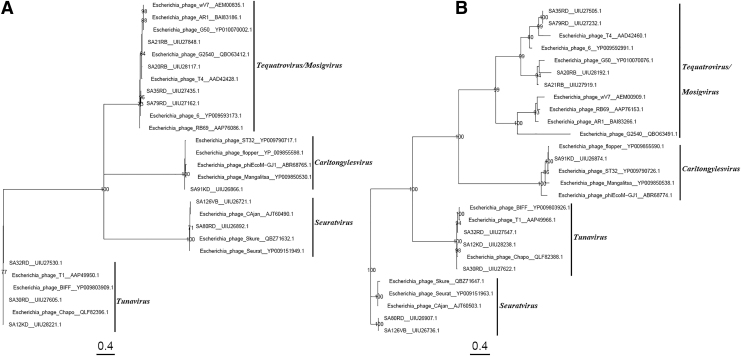
FIG. 2.
Phylogenetic tree was constructed using the maximum-likelihood method with 100 bootstrap replications to compare the capsid (A) and the tail fiber (B) proteins amino acid sequences of the non-O157 phages and other related Escherichia coli/STEC-infecting phage, chosen to represent the Tunavirus, Seuratvirus, Carltongylesvirus, Tequatrovirus, and Mosigvirus. The Genbank accession numbers of all amino sequences are as shown.