Fig 1. Overview of the m6A-SAC-seq protocol.
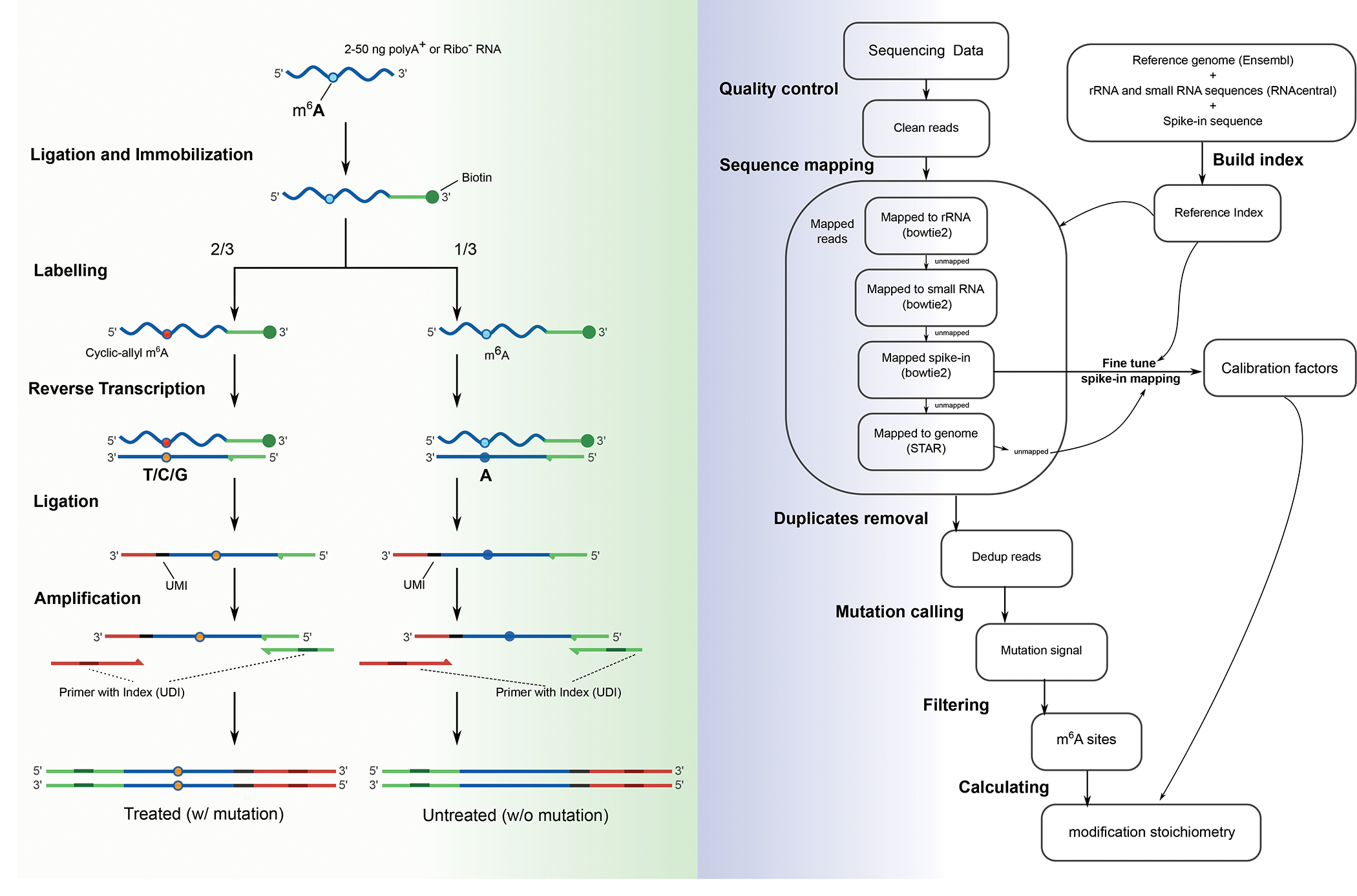
1a. Schematic representation of cDNA library preparation steps. 2–50 ng of poly-A enriched (polyA+) or ribosome RNA-depleted (Ribo−) RNAs are fragmented into short fragments (< 200 nt) and ligated to a 3’ biotinylated adaptor to facilitate on-beads immobilatization, then divided in a 2:1 ratio: 2/3 of the starting materials are labeled by MjDim1, whereas the remaining 1/3 serve as the untreated control. After reverse transcription, the cyclic allyl m6A sites (red dot) are converted to T/C/G mismatches (orange dot), whereas unconverted m6A sites (cyan dot) in the control group are read as A (blue dot). The obtained cDNA is ligated to another adapter carrying unique molecular identifiers (UMIs), then amplified by PCR with uniquely dual indexed primers (UDIs). The cDNA library is purified by size-selection so that fragments of more than 150 bp are recovered and they undergo NGS. 1b. Schematic representation of data analysis pipeline enabling estimation of the modification fraction (or mutation ratio) of the m6A sites.
