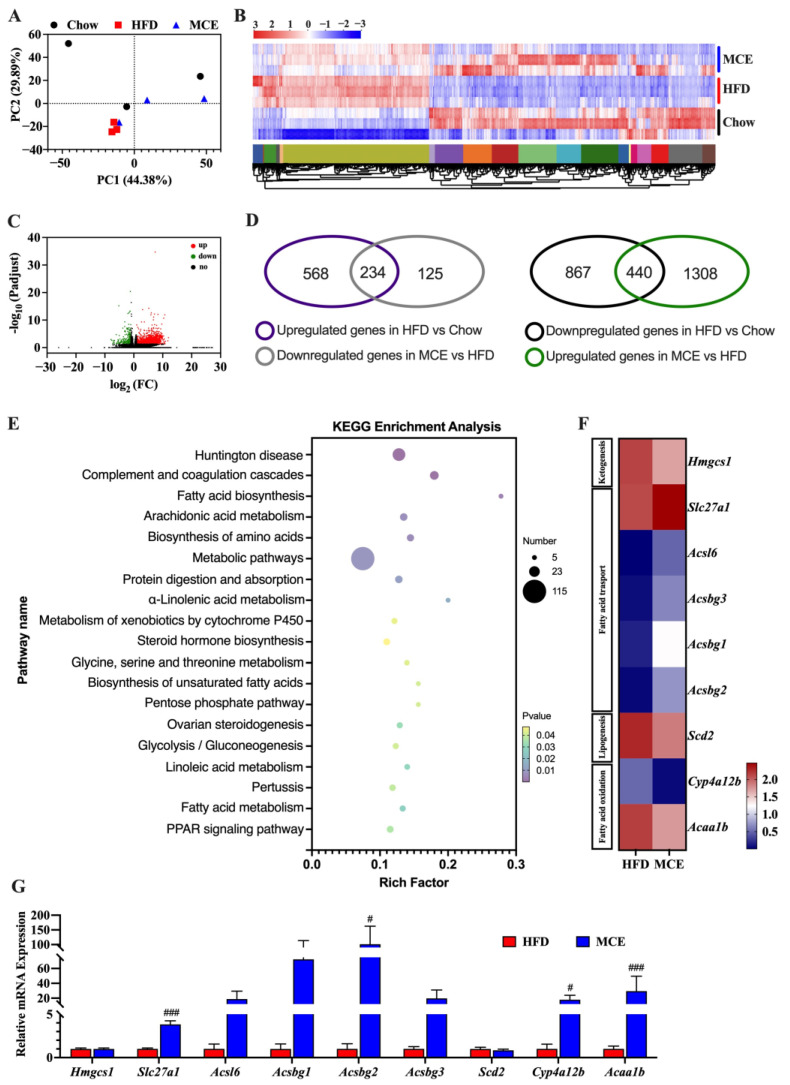
Figure 6.
MCE modifies the transcriptome profile of epididymal WAT in HFD-fed mice. (A) Principal component analysis of the epididymal WAT Transcriptome. (B) Hierarchical clustering of RNA-seq data from epididymal WAT samples is shown using a heatmap. (C) Volcano plots indicate the differential expression genes (DEGs) in the epididymal WAT between the HFD and MCE group. (D) Venn diagrams indicate the DEGs among chow, HFD, and MCE groups. (E) Enriched KEGG analysis of DEGs between HFD and MCE groups (p < 0.05). (F) Heatmap of the DEGs between HFD and MCE groups involved in the PPAR signaling pathway. n = 3. (G) mRNA levels of Hmgcs1, Scd2, Cyp4a12b, Acaa1b, Acsbg1, Acsbg2, Acsbg3, Slc27a1, and Acsl6 in epididymal WAT tested by RT-qPCR. n = 8. # p < 0.05 versus HFD group, ### p < 0.001 versus HFD group.