Table 1.
The binding affinity of the active compounds docked with the active site of the main protease of SARS-CoV-2 with a particular focus on the GLU 166, HIS 41, and CYS 145 residues.
| No. | Compound Name | Chemical Structure | Score (kcal/mol) |
Root-Mean-Square Deviation of Atomic Positions RMSD Range | Hydrogen Bonds (Number of Bonds/Number of Conformations), (Distance ≤ 4 Å) | Van der Waals (Number of Bonds/Number of Conformations), (Distance Range Å) |
|---|---|---|---|---|---|---|
| b | 2-(4a,8-Dimethyl-1,2,3,4,4a,5,6,7-octahydro-naphthalen-2-yl)-prop-2-en-1-ol |
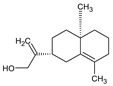
|
−5.6 | 27.83–29.77 | HIS 163 (1/1), SER 144 (1/1) | GLU 166 (19/1), (2.52–3.87) CYS 145 (1/1), (4.01) |
| d | 9beta-Acetoxy-3,5alpha,8-trimethyltricyclo[6.3.1.0(1,5)]dodec-3-ene |
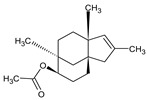
|
−5.6 | 33.48–36.10 | GLU 166 (1/1) | GLU 166 (5/2), (2.01–3.80) HIS 41 (6/2), (3.11–3.80) CYS 145 (4/2), (3.76–3.95) |
| c | 3-Oxatricyclo[20.8.0.0(7,16)]triaconta-1(22),7(16),9,13,23,29-hexaene |
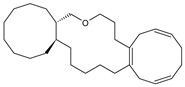
|
−7.8 | 27.51–30.19 | GLU 166 (1/1) | GLU 166 (6/1), (2.66–3.70) HIS 41 (2/1), (3.74–3.76) CYS 145 (2/1), (3.51 to 3.69) |
| d | 2(3H)-Benzofuranone, 6-ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-, [3S-(3.alpha.,3a.alpha.,6.alpha.,7.beta.,7a.beta.)]- |
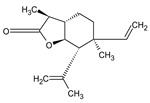
|
−5.6 | 0.00–0.00 | - | GLU 166 (8/1), (2.80–3.79) HIS 41 (5/1), (3.48–3.96) CYS 145 (1/1), (3.93) |
| e | Vanillosmin |
|
−6.4 | 30.75–33.33 | - | HIS 41 (4/1), (2.65–3.75), CYS 145 (1/1), (3.81) |
| f | Piperine |
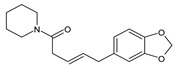
|
−6.0 | 30.24–32.01 | - | GLU 166 (5/1), (2.15–3.85) CYS 145 (2/1), (3.64–3.82) |
| g | Stigmasterol |
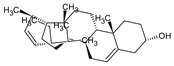
|
−6.8 | 31.13–33.56 | - | GLU 166 (7/1), (2.57–3.69) HIS 41 (5/1), (3.61–3.89) CYS 145 (3/1), (3.78–3.98) |
| h | Di(1,2,5-oxadiazolo)[3,4-b:3,4-E]pyrazine, 4,8-diacetyl- |
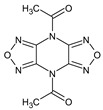
|
−6.5–−5.7 | 26.80–33.89 | PHE 140 (1/1), HIS 163 (1/1), HIS 164 (1/1) | GLU 166 (14/2), (2.49–3.62) HIS 41 (2/1), (2.96–3.69) CYS 145 (1/1), (3.62) |
| i | Cycloeucalenol acetate |
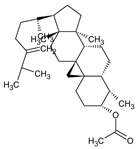
|
−7.0–6.4 | 25.29–34.14 | THR 26 (1/1) LYS 137 (1/1) MET 276 (1/1) |
GLU 166 (8/2), (2.67–3.81) HIS 41 (18/3), (2.87–3.90) CYS 145 (2/2), (3.73–3.77) |
| k | Strophanthidol |
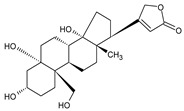
|
−7.2–6.7 | 0.00–3.98 | THR 26 (1/1) GLU 166 (3/2) GLN 198 (1/1) |
GLU 166 (27/3), (1.82–3.69) HIS 41 (8/3), (2.95–3.64) CYS 145 (2/2), (2.94–3.52) |
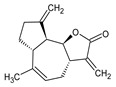
| l | Eudesma-5,11(13)-dien-8,12-olide |
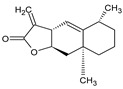
|
−6.1–5.9 | 29.56–33.61 | - | GLU 166 (2/1), (3.03–3.87) HIS 41(34/2), (1.70–3.96) CIS 145 (3/2), (3.77–3.88) |
| m | 2-Butenoic acid, 2-methyl-, 2-(acetyloxy)-1,1a,2,3,4,6,7,10,11,11a-decahydro-7,10-dihydroxy-1,1,3,6,9-pentamethyl-4a,7a-epoxy-5 |
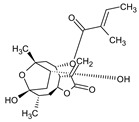
|
−6.6–6.1 | 29.19–32.40 | (ASN 142)2/2 GLU 166 |
GLU 166 (6/2), (1.99–3.20) HIS 41(14/2), (1.62–3.78) CIS 145 (6/2), (2.54–3.64) |
