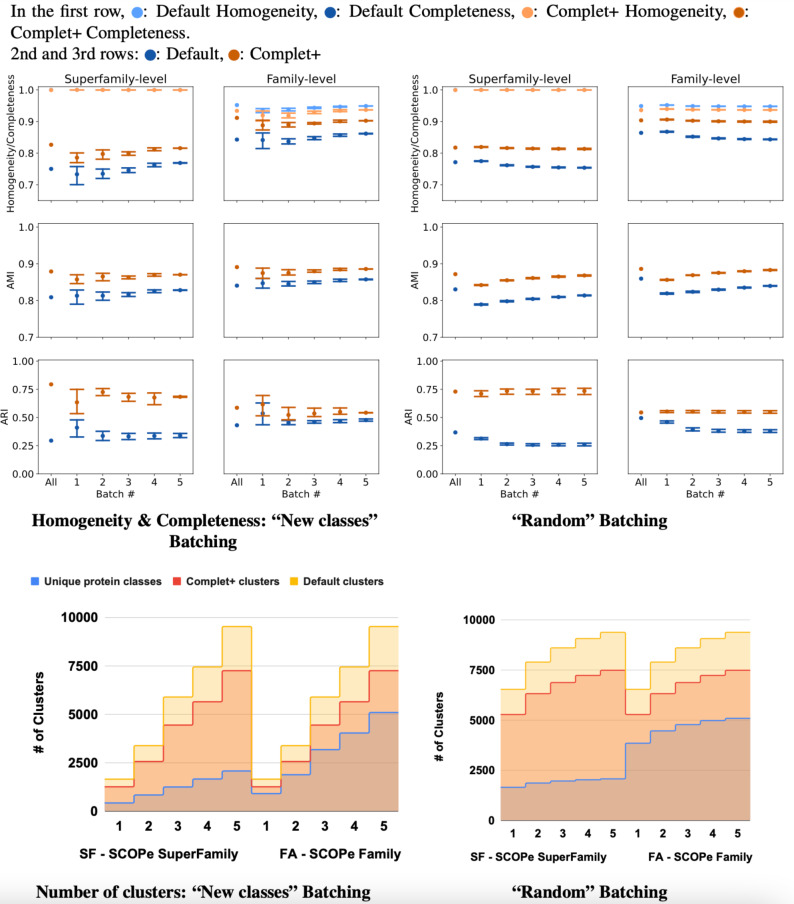
Figure 8. Homogeneity, completeness, AMI, and ARI of the superfamily- vs. family- level for both (A) “new classes” and (B) “random” test batching partitions (for five batches) for MMSeqs cluster and clusterupdate followed by Complet+.
The tick labeled “All” on the graphs represents clustering all sequences in one single batch. Overall, Complet+ increases MMSeqs2 completeness by substantially more than it reduces homogeneity relative to the default MMseqs2-generated clusters. Using Complet+ results in an increased AMI and ARI at both family and super-family levels. Also, we can see that discovery of new classes yield a large variance in performance as opposed to the base algorithm obtaining most classes in the first batch. The variance is due to the number of actual families or super-families (“true” clusters). (C & D) Number of true and predicted clusters for default MMseqs2 and Complet+. The number of true clusters is always lower than what Default MMseqs2 finds, and Complet+ is able to reduce them 10–20% by merging proteins that belong to the same family/superfamily.