Figure 2.
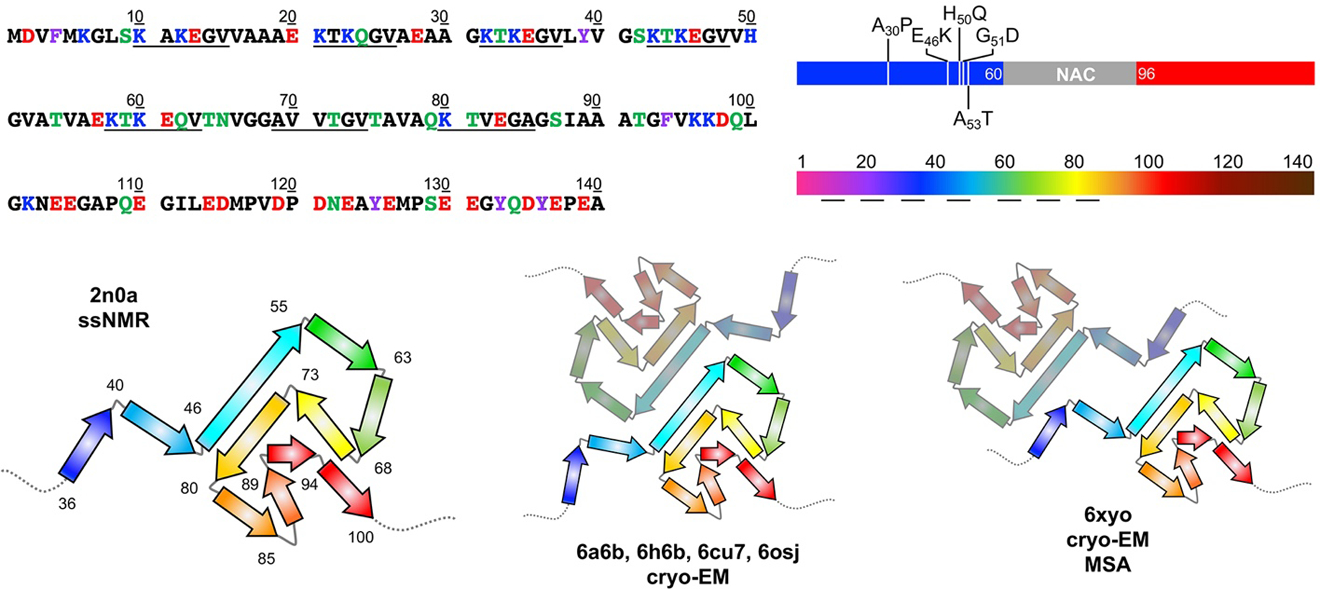
Top Left: αS sequence with amino acids colored by properties: hydrophobic (black), polar (green), aromatic (purple), acidic (red), or basic (blue). Imperfect repeats are underlined. Top Right: αS structure segments colored blue (N-terminal), grey (NAC), and red (C-terminal) as in Figures 1 and 3, with familial mutants indicated. Rainbow-colored by sequence number as in structural images below and Figures 4, 6, 7, and 8. Bottom: 2n0a ssNMR structures of αS fibrils viewed down the helical axis. The core region of residues 36–100 is shown in cartoon form with the residues at turns between β-stands noted. Cryo-EM structures of fibrils (6a6b, 6h6b, 6cu7, 6osj) showing a similar fold with a common protofibril packing motif observed in structures from several independent studies. A similar fold is also observed in fibrils from MSA patients (6xyo) with a different protofibril arrangement. PDB IDs are noted.
