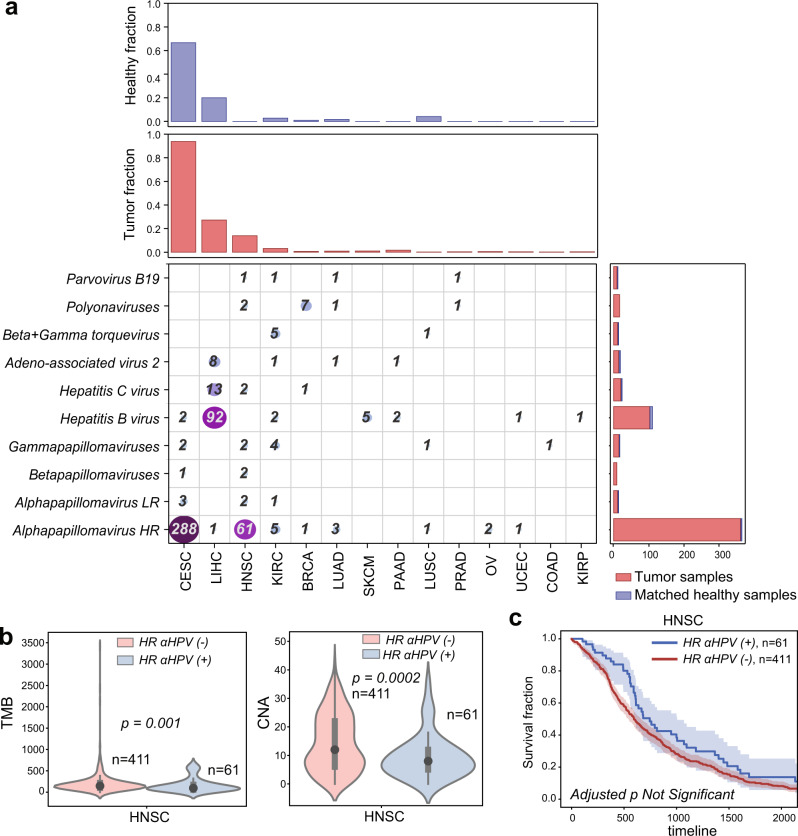
Fig. 2. Reference human viruses expressed in different tumor types.
a Heatmap showing the total number of virus-positive samples identified from RNA sequencing in different tumor tissues. The top panels show the fraction of tumor and non-cancer samples in which viruses were identified. The right panels show the number of viruses found in tumor and non-cancer samples. b Violin plots comparing the tumor mutation burden (TMB) and the number of chromosome-level copy number alteration (CNA) between HNSC patients where expression of high-risk alpha papillomaviruses was detected vs those patients where expression of high-risk alpha papillomaviruses was not detected. Black dots represent the medians, and the boundaries of the violin plots refer to the maximum and minimum values, respectively. Two-sided Wilcoxon rank-sum p value is reported. c Kaplan–Meier curves comparing the survival rates between HNSC patients where the expression of high-risk alpha papillomaviruses was detected (blue curve) vs those where the expression of high-risk alpha papillomaviruses was not detected (red curve). The FDR-adjusted two-sided log-rank p value is not significant (Supplementary Table 1). For Kaplan–Meier curves, shaded areas represent the confidence interval of survival. Source data are provided as a Source Data file 2.