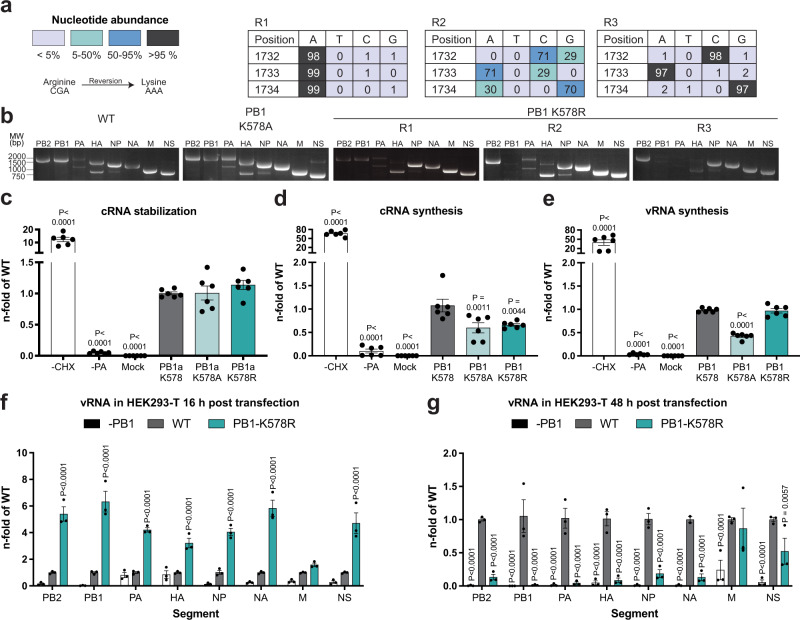
Fig. 5. Mutation of PB1-K578 affects cRNP stabilization and vRNA synthesis.
a, b Detection of packaged vRNA segments isolated from released virus particles in the supernatants of the rescue experiments at 48 h p.t. vRNA segments were amplified with segment-specific primers, separated on 2% agarose gel (b). Gels from all three replicates of K578R and representative gels of three replicates (R) with consistent results are shown for WT and PB1-K578A. vRNA was analyzed using deep sequencing. Percentage of nucleotides encoded at position 1732–34 encoding for K578 (a). c–e Analysis of cRNA stabilization, cRNA and vRNA synthesis. HEK293-T cells were transfected with plasmids expressing NP, PB2, PA, inactive PB1 [PB1a] (D445A/D446A62,100; c) or active PB1 (d, e). At 24 h p.t., cells were infected with WT WSN virus (MOI: 5) and cultured in infection medium with cycloheximide (CHX). At 6 h p.i. cRNA (c, d) or vRNA levels (e) of the NA segment were assessed using strand-specific RT-PCR and are shown as mean n-fold of WT (±SEM), n = 6 independent biological replicates. P values < 0.05 compared to WT from Dunnett’s multiple comparison one-way ANOVA-test are indicated. f, g Accumulation of vRNA segments in HEK293-T cells co-transfected with pHW2000 plasmids at the indicated time points. vRNA levels were quantified using segment-specific primers for RT-PCR and presented as the mean n-fold of WT (±SEM), n = 3 independent biological replicates. Cellular GAPDH mRNA levels were used as housekeeping control. P values < 0.05 compared to WT from Šidák’s corrected multiple comparison two-way ANOVA test are indicated. Source data are provided as a Source data file.