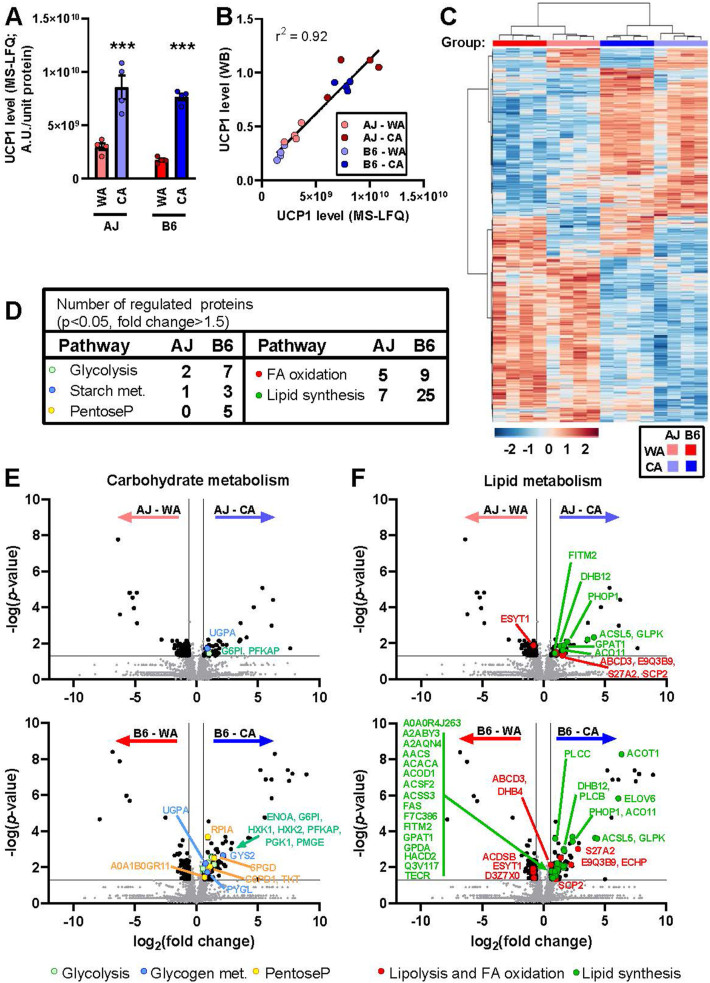
Figure 5.
Acclimation temperature dominates over the mouse strain in the effects on iBAT proteome. Analyses of iBAT proteome of AJ and B6 mice acclimated to a thermoneutral temperature (30 °C; WA) or to cold (6 °C; CA) was performed using mass-spectrometry label-free quantification (MS-LFQ; n = 4). (A) Specific UCP1 content in iBAT homogenate; data are means ± SEM; ∗∗∗p < 0.001 vs. the WA mice (two-way ANOVA and Tukey's multiple comparison test). (B) Comparison of UCP1 quantification using MS-LFQ and Western blotting (WB); analysis performed as in Figure 2B. (C) Hierarchical clustering of proteins based on MS-LFQ intensity. Each column represents an individual animal (experimental group is indicated by colour code above the column), each row represents an individual protein. Only proteins differing significantly among the experimental groups (one-way ANOVA) were considered (i.e. 570 out of 3501 proteins detected). Both mice and proteins were automatically clustered using MetaboAnalyst (v 4.0 and 5.0) software [70] as indicated by dendrograms above and left of the plot. For the list of proteins, see S2 Data (sheet “BAT_ANOVA”). Hue represents the autoscaled t-test/ANOVA score. (D, E, F) Effect of temperature on the abundance of individual proteins in AJ and B6 mice. Total number of proteins/enzymes of selected metabolic pathways affected by cold in AJ and B6 mice (D). Volcano plots showing effects of temperature on proteins involved in glucose (E) and lipid metabolism (F). For the background data see S2 Data). For the effect of the strain on the above proteins, see S3 Fig. For the Volcano plot analysis of the quantitative composition of protein subunits of mitochondrial oxidative phosphorylation system, see S4 Fig. Volcano plots to demonstrate the difference in quantitative proteome composition between the WA- and the CA-mice, based on all 3501 proteins detected; S2 and S3 Data); plotted separately for AJ and B6 mice (upper and lower panels, respectively). Significantly regulated proteins (i.e. p-value <0.05; fold change >1.5) were (i) indicated by black dots (in AJ-WA vs. AJ-CA comparison: 48 proteins upregulated by CA and 85 proteins downregulated by CA; in B6-WA vs. B6-CA comparison: 140 proteins upregulated by CA and 93 proteins downregulated by CA; see S3 Data); (ii) ascribed to major metabolic pathways using KEGG database (see S3 Data); and (iii) color-coded according their involvement in carbohydrate (E) or lipid (F) metabolism. For the protein codes, see the entry name of UniProt database (used here without the name of the organism, e.g. UCP1 is originally UCP1_MOUSE; see S3 Data). (A, B) Values for each data point can be found in S1 Data.