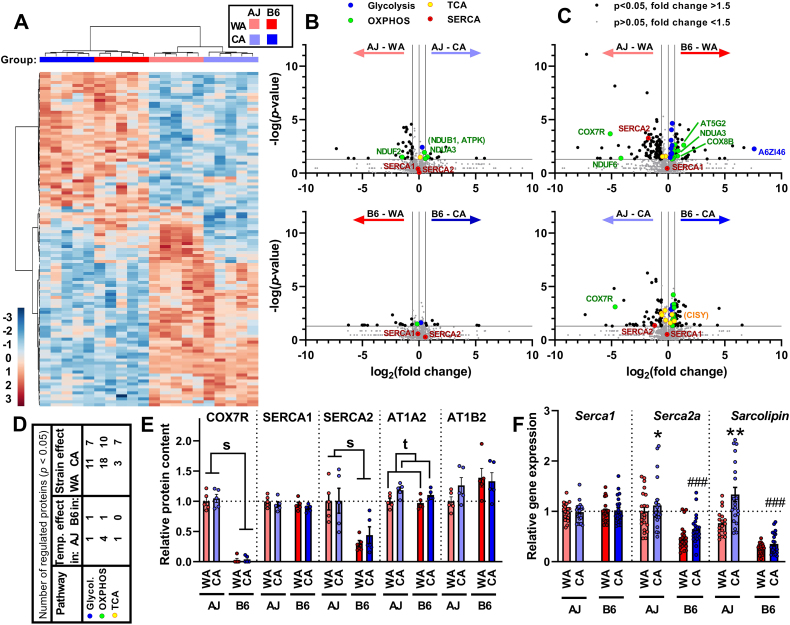
Figure 7.
Acclimation temperature exerts a weaker effect compared with the mouse strain on skeletal muscle proteome. Analyses of gastrocnemius muscle proteome of AJ and B6 mice acclimated to a thermoneutral temperature (30 °C; WA) or to cold (6 °C; CA) was performed using mass-spectrometry label-free quantification (MS-LFQ; n = 4). (A) Hierarchical clustering of proteins based on MS-LFQ intensity. Each column represents an individual animal (experimental group is indicated by color code above the column), each row represents an individual protein. Only proteins differing significantly among the experimental groups (one-way ANOVA) were considered (i.e. 112 out of 1781 proteins detected in total). Both proteins and mice were automatically clustered using MetaboAnalyst (v 4.0 and 5.0) software [70] as indicated by dendrograms above and left of the plot. For the list of proteins, see S2 Data (sheet “GASTRO_ANOVA”). Hue represents the autoscaled t-test/ANOVA score. (B, C, D) Differences in proteome among the experimental groups. Volcano plots to demonstrate the difference in quantitative proteome composition between the WA- and the CA-mice (B) and between the AJ and B6 mice (C), based on all 1771 proteins detected; S2 and S3 Data). Significantly regulated proteins (i.e. raw p-value <0.05; fold change >1.5) were (i) indicated by black dots (in AJ-WA vs. AJ-CA comparison: 12 proteins upregulated by CA and 32 proteins downregulated by CA; in B6-WA vs. B6-CA comparison: 8 proteins upregulated by CA and 17 proteins downregulated by CA; in AJ-WA vs. B6-WA comparison: 100 proteins upregulated in AJ and 39 proteins upregulated in B6; in AJ-CA vs. B6-CA comparison: 56 proteins upregulated in AJ and 30 proteins upregulated in B6 see S3 Data); (ii) ascribed to major metabolic pathways using KEGG database (see S3 Data). The selected proteins are labelled by protein codes (for the protein codes, see the entry name of UniProt database - used here without the name of the organism, e.g. COX7R is originally COX7R_MOUSE; see S3 Data). Significantly regulated proteins mentioned in text but with fold change <1.5 are labelled by code in parentheses.). (D) Number of differentially regulated proteins/enzymes engaged in selected metabolic pathways (see B and C) in mice of both strains (see S3 Data). (E) Levels of selected proteins (see B and C and S3 Data; n = 4). (F) Expression of selected genes (measured using qPCR; data were normalized to a geometric mean of 2 housekeeping genes, see Materials and Methods). Data combined from 3 independent experiments; n = 18–21; data are means ± SEM; ∗p < 0.05 and ∗∗p < 0.01 vs. the WA mice; ###p < 0.001 vs. the AJ mice (two-way ANOVA and Tukey's multiple comparison test). (E, F) Values for each data point can be found in S1 Data.