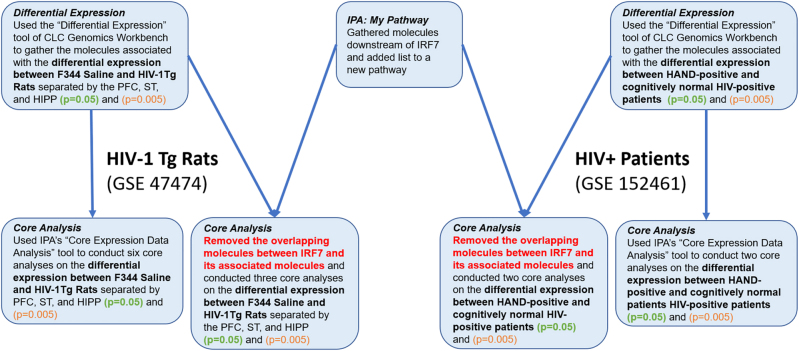
Figure 2:
The flow of steps involved in using the bioinformatics and data collection tools offered by QIAGEN to simulate the complete knockout of IRF7 and its associated molecules in F344 rats and HIV-positive individuals. First, the “Differential Expression” tool of CLC genomics workbench was used to determine the differential expression between F344 Saline and HIV-1Tg rats and between HAND-positive and cognitively normal HIV-positive individuals. Then, the a “Canonical Pathway Analysis” was performed on the molecular datasets. Next, the “My Pathway” tool of IPA was used to identify the 455 molecules associated with IRF7. The overlapping molecules between the molecules associated with IRF7 and the molecules in the differential expression datasets were removed. Finally, the “Canonical Pathway Analysis” was performed on the datasets without the 455 molecules associated with IRF7.