Extended Data Fig. 1. Human and mouse breast cancers that become senescent-like post chemotherapy enrich the same immune modulatory programs.
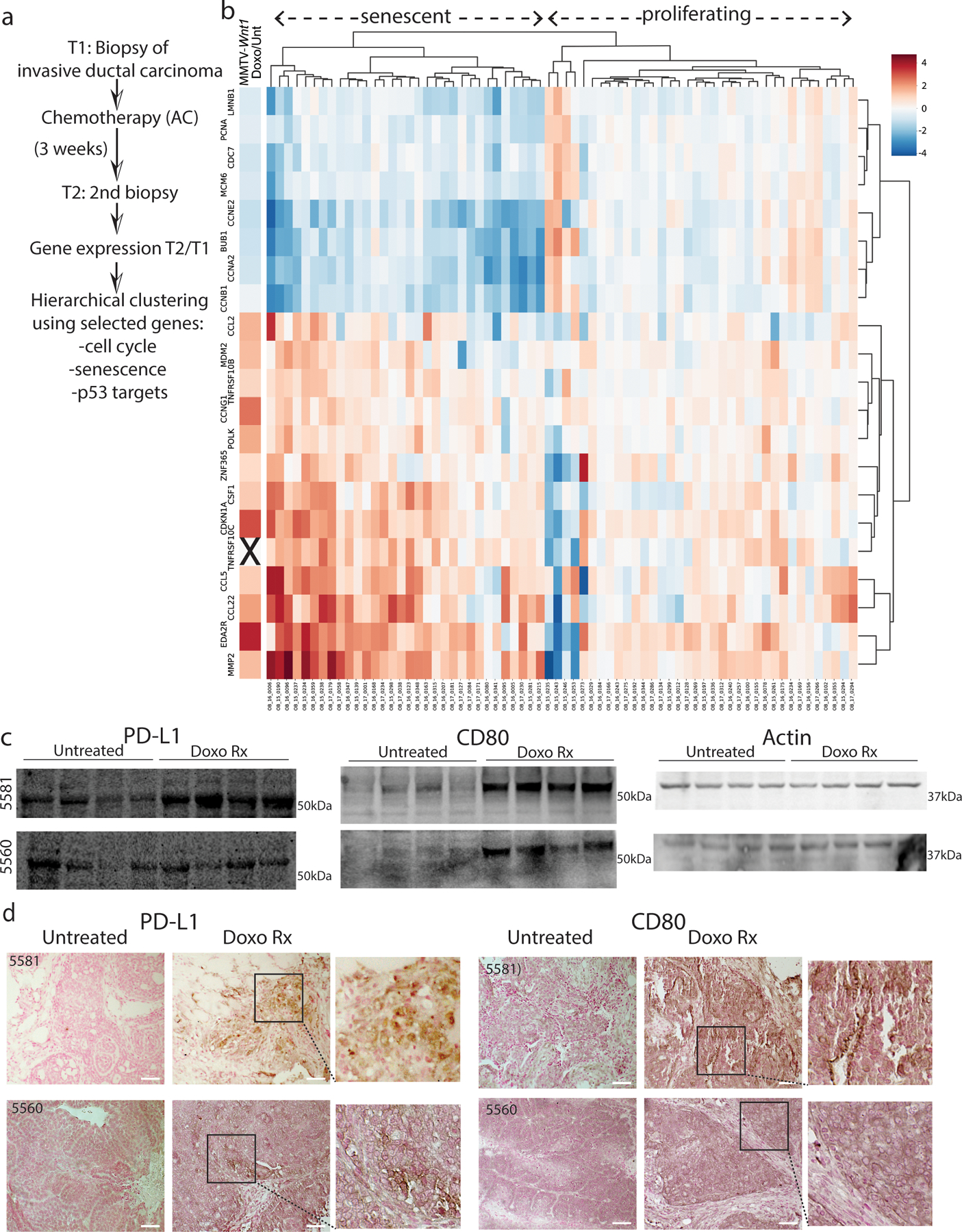
A) Summary of breast cancer patient treatment scheme and RNA-seq analysis. B) Hierarchical clustering of an RNA seq dataset19 was performed on NAC post/pre fold-change values for each tumor, using cell cycle genes (PCNA, CDC7, MCM6, CCNE2, BUB1, CCNA2, CCNB1), senescence/ SASP genes (LMNB1, CCL2, TNFRSF10B, CSF1, CCL5, CCL22, MMP2) and p53 targets (CDKN1A, CCNG1 POLK, ZNF365, EDA2R) as justified in Supplemental Table 1. The leftmost column represents treated/untreated fold change in expression of each gene, derived from RNA seq of spontaneous MMTV-Wnt1 tumors. These tumors are well characterized to undergo senescence post treatment4–6 but were not used in the clustering. C-D) Two additional p53 wild-type MMTV-Wnt1 tumors (5581, 5560) were each transplanted and mice were treated and harvested as in Fig. 1D. Indicated immunoblots were performed. D) Formalin fixed paraffin embedded (FFPE) tumor sections from treated or untreated mice as in Fig. 1E were IHC stained for PD-L1 or CD80 and counter stained with nuclear fast red. Scale bar=50µm. Western blots and IHC were repeated at least twice and representative data are shown.
