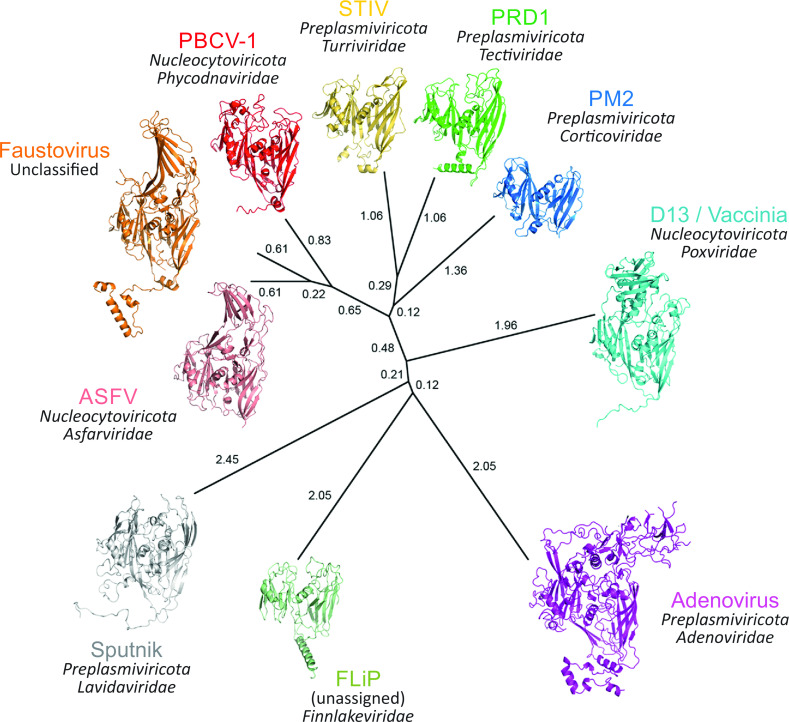
Fig 2. Structure-based dendrogram of capsid proteins of members of the kingdom Bamfordvirae.
Structure-based phylogenetic tree inferred from major capsid protein (MCP) structures of the members of the kingdom Bamfordvirae in the Varidnaviria realm. Members of Bamfordvirae encode a vertical double-jelly roll fold MCP, which is the hallmark protein of this group of viruses. Next to each MCP structure are the virus name (top), the phylum (middle), and family (bottom), with “Faustovirus” not yet officially classified and Finnlakeviridae not yet assigned to any higher taxon. The evolutionary distances across the depicted members of the originally called PRD1-adenovirus viral lineage [67] were calculated with the Homologous Structure Finder software [50] and depicted with PHYLIP (https://evolution.genetics.washington.edu/phylip.html); the evolutionary distances are shown next to each branch. The protein data bank identifiers (PDBid) for the structures are as follows: PRD1: PDBid 1HX6; PBCV-1: 1M3Y; adenovirus: 1P2Z; STIV: 2BBD; Vaccinia D13: 2YGB; Sputnik: 3J26; Faustovirus: 5J7O; FLiP: 5OAC; ASFV p72: 6KU9; PM2: 2W0C. Adapted from [62].