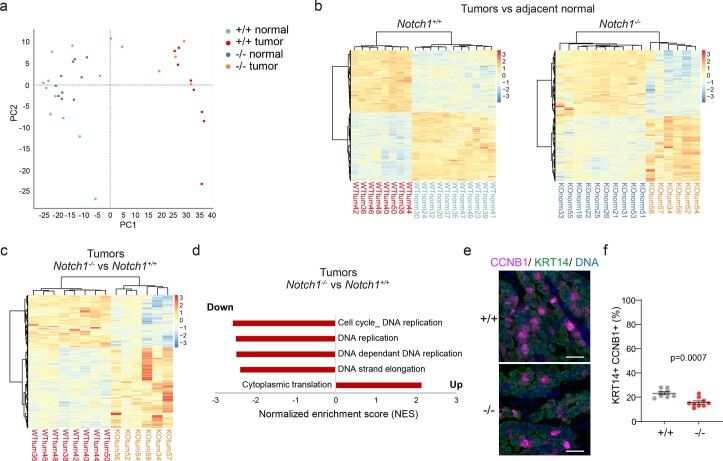
Extended Data Fig. 10. Transcriptomic characterization and cellular phenotype of tumors from Notch1+/+ and Notch1−/− esophagus.
a-d. RNA-seq analysis of Notch1+/+ and Notch1−/− esophageal tissue and tumors, see Fig. 8a. a. Principal component analysis (PCA) plot showing in two dimensions all biological replicates from Notch1+/+ normal epithelium (n = 11 biopsies from 7 mice, green), tumors from Notch1+/+ epithelium (n = 8 tumors from 4 mice, red), Notch1−/− normal epithelium (n = 10 biopsies from 7 mice, blue) and tumors from Notch1−/− epithelium (n = 6 tumors from 5 mice, orange). Dotted lines indicate the origin of the axes. b. Hierarchical clustering and heat map showing differentially expressed genes between tumors and adjacent normal tissue, in Notch1+/+ mice (left) and Notch1−/− mice (right). c. Hierarchical clustering and heat map showing differentially expressed genes between tumors from Notch1+/+ and Notch1−/− esophagus. d. Gene Set Enrichment Analysis (GSEA) of tumors from Notch1−/− vs Notch1+/+esophagus. Normalized enrichment score (NES) of altered Biological Process gene sets in tumors. False discovery rate, FDR q-value<0.05. e. Representative images of n = 8 tumors from Notch1+/+ and n = 9 tumors from Notch1−/− esophagus. KRT14 (green), CCNB1 (magenta). DNA, blue. Scale bars, 30 µm. f. Percentage of CCNB1 positive; KRT14 expressing keratinocytes within tumors from Notch1+/+ and Notch1−/− esophagus. Mean ± SEM, each dot represents a tumor, +/+: n = 8 tumors from 4 mice; −/−: n = 9 tumors from 7 mice. Two-tailed unpaired Student’s t-test. See Supplementary Tables 24 and 27–29.