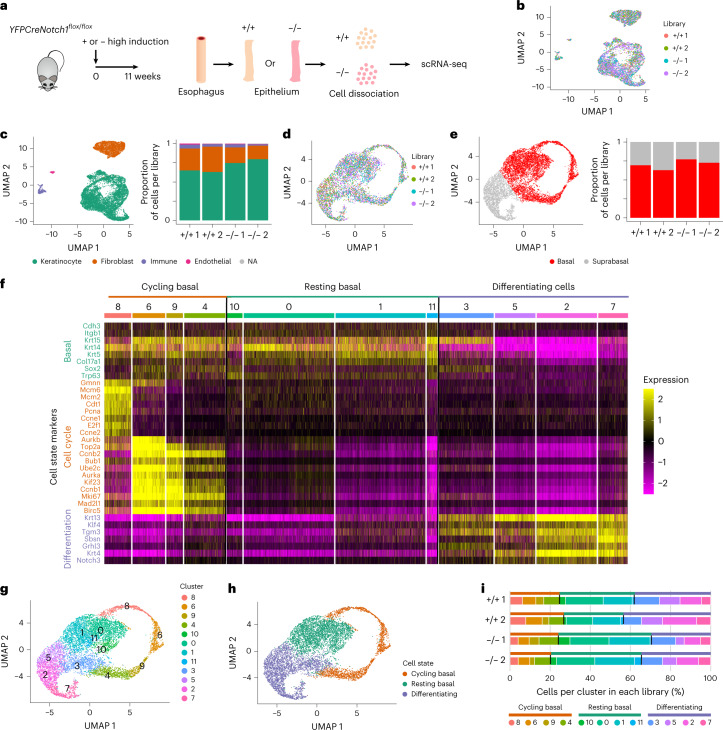
Fig. 4. Notch1 loss does not alter tissue composition or cell dynamics.
a, YFPCreNotch1flox/flox mice were highly induced and aged for 11 weeks, allowing the mutant cells to completely occupy the esophageal epithelium. Controls were uninduced YFPCreNotch1flox/flox mice (+/+). Esophageal epithelium was dissociated and sequenced. b, UMAP plot shows an overlay of 1,500 cells from each library (n = 2 mice per genotype; +/+1, n = 2,454; +/+2, n = 3,194; −/−1, n = 1,929; −/−2, n = 5,534). c, Left, UMAP plot showing cell types identified via scRNA-seq. Right, stacked bar chart shows the proportion of cell types per library. NA, not available. d, UMAP plot shows an overlay of 1,400 cells annotated as keratinocytes from each library (+/+1, n = 1,555; +/+2, n = 1,932; −/−1, n = 1,403; −/−2, n = 3,919). Milo test shows no significant difference in local cell density through UMAP space (Supplementary Note). e, Left, UMAP plot of keratinocytes. Right, stacked bar chart shows the estimated proportion of keratinocytes per library belonging to the basal or suprabasal layers (Supplementary Note). f, Heat map showing Seurat processed expression values in the keratinocyte population for representative marker genes of basal cells, cell cycle, and differentiation for the 11 clusters shown in g (marker list from ref. 32). Clusters are grouped in three different cell states: cycling basal, resting basal and differentiating cells. g, UMAP plot of keratinocytes representing cell clusters based on Seurat analysis pipeline via the Leiden algorithm. h, UMAP plot of keratinocytes showing cycling basal (orange), resting basal (green) and differentiating (purple) cell states based on clusters and differentiation markers analysis performed in f and g. i, Stacked bar charts show the proportion of keratinocytes per cell state (upper bar) and per cluster (lower bar) in each library. See Supplementary Table 16.