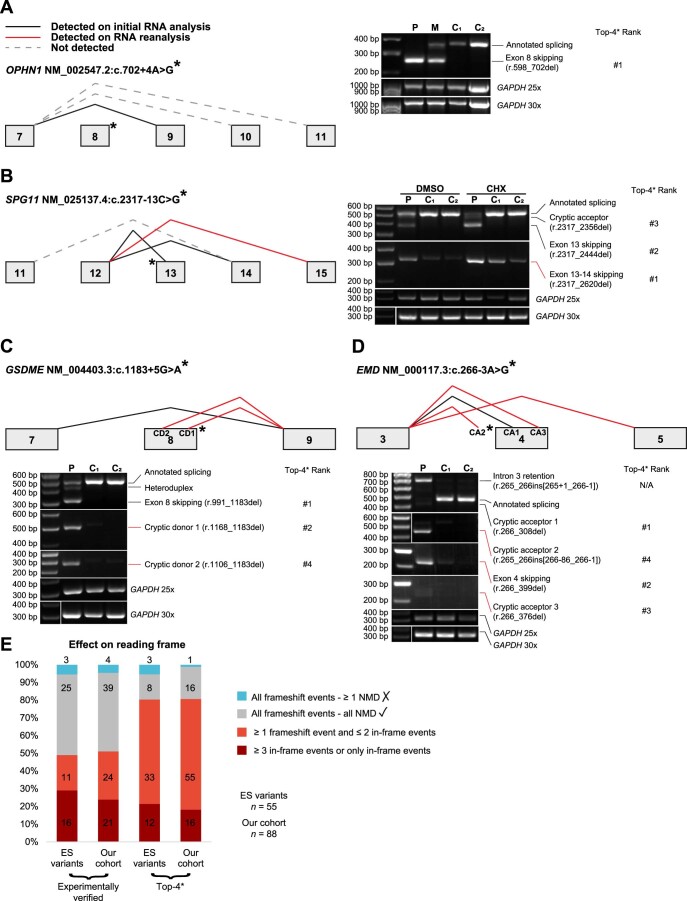
Extended Data Fig. 8. RNA re-analysis to check for undetected 300K-RNA Top-4 mis-splicing events.
Black lines: mis-splicing identified during initial RNA analysis7. Red lines: Top-4* events detected upon re-analysis. Gray lines: Top-4* events undetected upon re-analysis. a) No additional Top-4 events were identified for OPHN1 c.702 + 4 A > G. b) For SPG11 c.2317-13 C > G, Top-1* exon 13 + 14 skipping was detected on re-analysis (missed initially by RT-PCR due to primer positioning and by RNA-seq due to low read depth and NMD), but Top-4* event, exon 12 + 13 skipping, was not detected. c) RT-PCR using primers specific for two exonic cryptic donors shows their variant-associated increased use for GSDME c.1183 + 5 G > A. These rare events were missed during initial RNA analysis due to PCR biases and challenges resolving Sanger sequencing chromatograms due to heteroduplex formation. d) RT-PCR identifies rare use of two cryptic acceptors and exon 4 skipping associated with EMD c.266-3 A > G missed during initial RNA analysis due to PCR biases and heteroduplex formation. e) 49% (27/55) essential splice-site variants (ES) across the three variant datasets induce ≥ 1 in-frame events, with a similar proportion of 51% (45/88) in our overall cohort. Use of Intron Retention (IR) and Top-4* as proxy for a prediction of variant-induced mis-splicing increases the relative number of ES variants with ≥1 in-frame event to 80% (45/55) and to 81% (71/88) for our overall cohort. P = proband, C1 = control 1, C2 = control 2, DMSO = dimethyl sulfoxide, CHX = cycloheximide, CD1 = cryptic donor 1, CD2 = cryptic donor 2, CA1 = cryptic acceptor 1, CA2 = cryptic acceptor 2, CA3 = cryptic acceptor 3, ES = essential splice-site, NMD = nonsense mediated decay.