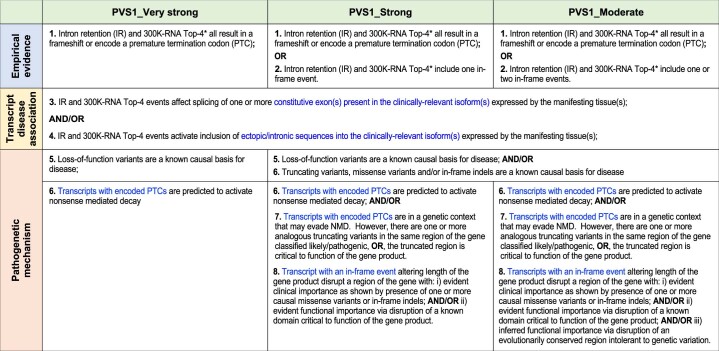
Extended Data Fig. 10. Draft Guidelines for potential use of empirical evidence from 300K-RNA to assist application of the PVS1 criterion for essential splice-site variants based on probable mis-splicing outcomes.
(being assessed in a clinical evaluation trial by the Australian Consortium for RNA Diagnostics (SpliceACORD)). We recommend pathology consideration of Intron Retention (IR) and 300K-RNA Top-4* for all disease relevant transcript(s). PVS1 levels of evidence are influenced by the collective nature of probable induced mis-splicing, relative to evidence supporting null outcomes for the encoded gene product. For use of PVS1 at a Very Strong evidence level, IR and Top-4* events should all be consistent with null outcomes. PVS1 applied at Strong or Moderate should be considered when IR and Top-4* events include one or two in-frame events, adjusting the evidence weighting according to; the number and nature of in-frame events, known clinical relevance of the affected region of disease relevant transcript(s), and the established pathogenetic mechanism(s) associated with a given gene and disorder. We favor weighting of known clinical relevance, biological function or evolutionary conservation of the disrupted gene region, over relative length of the in-frame disruption. We recommend additional consideration of abnormal or alternative transcription initiation or termination for first intron and last intron variants, respectively. We recommend use of the PM4 criterion for essential splice-site variants when IR and Top-4* events include three or more in-frame events.