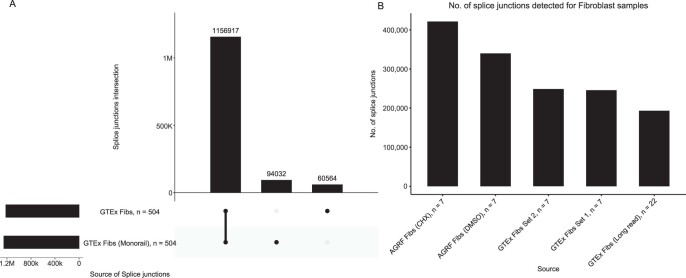
Extended Data Fig. 4.
a) Upset plot showing splice-junctions concordantly and uniquely detected by GTEx and Monorail processing pipelines for 504 fibroblast specimens. b) Upset plot showing GTEx V9 long-read RNA-Seq (7 M mean read depth, 740 nt average length) for 22 fibroblast specimens identifies 15% (180 K/1.16 M) of all fibroblast SJ detected in 504 GTEx v8 fibroblast samples (75 bp paired reads, 80 M depth, Poly A enrichment). In-house RNA-Seq data from 7 fibroblast specimens (150 bp paired reads, 100–200 M depth, rRNA depleted total RNA) subject to cycloheximide (CHX) treatment identifies 36% (420 K/1.16 M) of all SJ, substantially more that DMSO treated specimens (340 K/1.16 M) or in two randomized sets of 7 fibroblast specimens from GTEx v8 (245 K/1.16 M). Splice junctions present in GTEx v9 long read RNA-Seq data was reverse-engineered from the transcript count information generated using FLAIR27. AGRF, Australian Genome Research Facility; Fibs, fibroblasts.