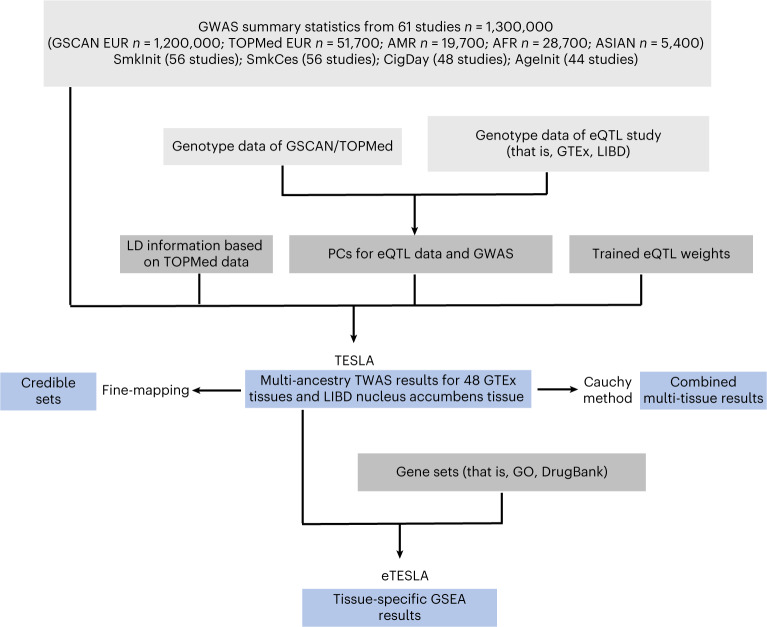
Fig. 1. Schematic description of the TESLA method.
TESLA uses meta-regression to model phenotypic effect estimates as functions of the PCs of genome-wide allele frequencies from each cohort. For a given gene expression prediction model generated from an eQTL dataset, we use TESLA to more accurately estimate phenotypic effects, then use them to perform TWASs and attain optimal power. We also performed fine mapping and enrichment analysis using the TESLA results (which we call eTESLA).