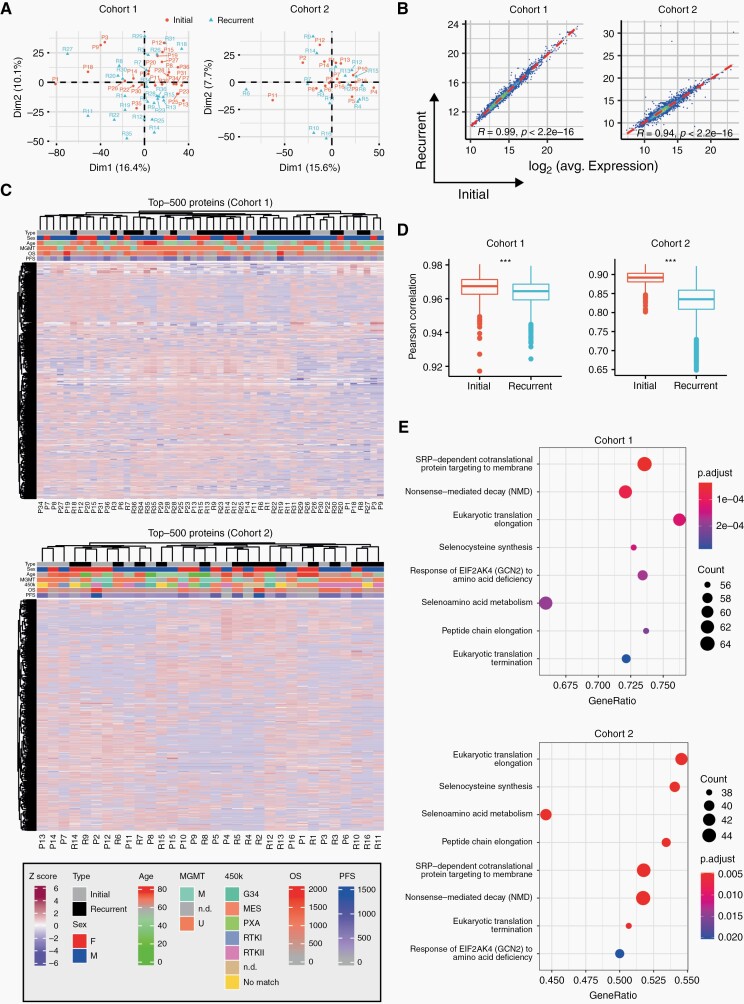
Fig. 2.
Quantitative proteomic landscapes of newly diagnosed and recurrent glioblastoma are similar. (A) Principal component analysis based on global proteomic data for cohort 1 (left) and cohort 2 (right). (B) Pearson correlation between initial and recurrent glioblastoma samples based on averaged protein abundance. (C) Hierarchical clustering based on the 500 most variable proteins for cohort 1 (top) and 2 (bottom). MGMT, methylation promoter methylation status (U, unmethylated, M, methylated, n.d., not determined). OS, overall survival; PFS, progression-free survival. Sex: f, female; m, male. 450k: DNA methylation class annotation. (D) Pearson correlations of different patient samples based on the global proteome calculated individually within initial and recurrent samples. P values were derived from a two-sided Welch test. (E) Significantly enriched Reactome pathways identified by gene set enrichment analysis. Only the least redundant pathways shared between both cohorts are shown (top: cohort 1, bottom: cohort 2; set size: 50-150; colors: FDR-adjusted P values; dot size: number of detected proteins per gene set, x-axis: enrichment ratio).