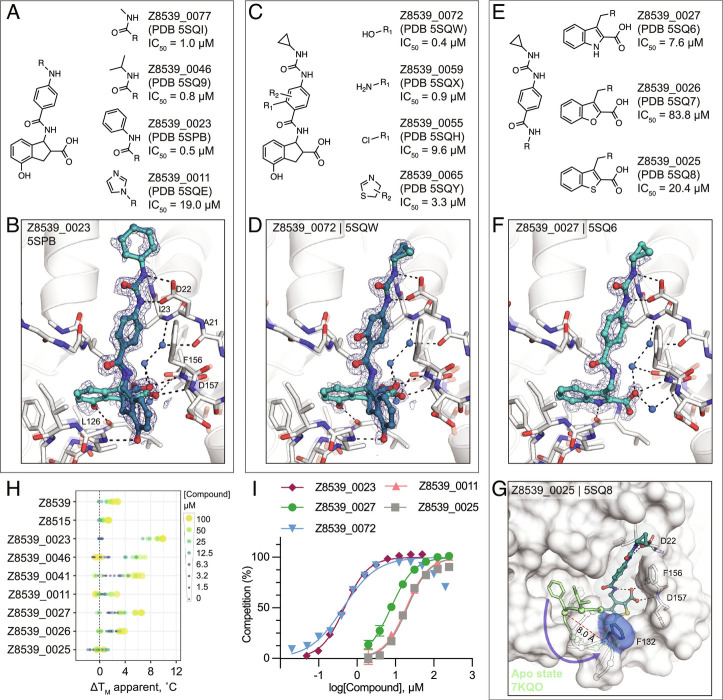
Fig. 3.
Structure-based optimization of Z8539. A) Modifications of the cyclopropyl-phenylurea group. B) X-ray crystal structure of Mac1 bound to Z8539_0023. The PanDDA event map is shown around the ligand (blue mesh contoured at 2 σ). C) Modifications of the central benzene. D) X-ray crystal structure of Mac1 bound to Z8539_0072. E) Modifications of the indane group. F) X-ray crystal structure of Mac1 bound to Z8539_0027. G) X-ray crystal structure of Mac1 bound to Z8539_0025. The Gly130-Phe132 loop is aligned to the apo-state conformation in green (PDB 7KQO). The Z8539_0025-Mac1 structure is shown with a transparent white surface. H) DSF-derived temperature upshifts. Data are presented for three technical replicates. I) HTRF-based peptide displacement dose–response curves. Data are presented as the mean ± SEM of at least two technical replicates.