Significance
Despite their central role in health and disease, the cellular mechanisms underlying how plasma cells persist while producing large quantities of antibodies are still poorly understood. In this paper, we describe that plasma cell survival is under the strict dependency of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) molecule Sec22b. We showed that this molecule is a key regulator of endoplasmic reticulum and mitochondrial structure in plasma cells, and in its absence, the humoral immune response is abrogated. Our findings have important implications for our understanding of antibody-mediated immunity but also open avenues for targeting plasma cells in pathological contexts.
Keywords: plasma cell, SNARE, antibody, endoplasmic reticulum, mitochondria
Abstract
Despite the essential role of plasma cells in health and disease, the cellular mechanisms controlling their survival and secretory capacity are still poorly understood. Here, we identified the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) Sec22b as a unique and critical regulator of plasma cell maintenance and function. In the absence of Sec22b, plasma cells were hardly detectable and serum antibody titers were dramatically reduced. Accordingly, Sec22b-deficient mice fail to mount a protective immune response. At the mechanistic level, we demonstrated that Sec22b contributes to efficient antibody secretion and is a central regulator of plasma cell maintenance through the regulation of their transcriptional identity and of the morphology of the endoplasmic reticulum and mitochondria. Altogether, our results unveil an essential and nonredundant role for Sec22b as a regulator of plasma cell fitness and of the humoral immune response.
Plasma cells (PCs) are the cellular source of humoral immunity via the long-term secretion of large quantities of antibodies that provide protection against reinfection. These cells can also contribute to diseases including plasmacytomas as well as antibody-mediated autoimmune and inflammatory pathologies. However, the therapeutic arsenal to target PCs is still very limited. Despite the essential role of these cells in health and disease, the cellular mechanisms controlling their secretory function and their survival are poorly understood. Closing this knowledge gap is thus of paramount importance for designing approaches to target this cell type.
During the transition from B cell to PC, the cell is reprogrammed to produce and secrete around 102 to 103 antibodies per second (1). To accommodate this large protein load, PCs expand their endoplasmic reticulum (ER) and adapt to tolerate the extra stress induced via upregulation of the Ire1α/Xbp1 branch of the unfolded protein response (UPR) (2–4). Early works from the 70s report that antibody secretion happens via the conventional constitutive exocytosis pathway meaning that they are not prestocked in granules but secreted as they are produced and trafficked from the ER to the Golgi apparatus where they are glycosylated (5–7).
Here, through a proteomics analysis, we identified the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) Sec22b as an interesting molecule up-regulated in mouse and human PCs. Members of the SNARE family are essential for the intracellular transport and fusion of protein cargoes between organelles. They are involved in both regulated and constitutive exocytosis and form a large family composed of different subtypes (Qa-, Qb-, Qc-, and R-SNAREs) that interact to form a ternary SNARE complex allowing membrane fusion between vesicles and organelles (8). Distinct sets of SNAREs are expressed on the different organelles and can also be cell type specific (9). On top of vesicular transport, Sec22b has other noncanonical functions, some of which could be highly relevant for PC biology, including ER branching, nuclear shuttling of transcription factors, and plasma membrane expansion (10–13). Using a conditional knock-out (KO) mouse model, we demonstrated that Sec22b is essential for PC maintenance and effective humoral response after vaccination and infection. At the mechanistic level, we showed that Sec22b governs expression of genes involved in not only cell cycle but also mitochondrial function and ER structure. Accordingly, Sec22b-deficient PCs displayed a dramatically altered ER network, reduced contact between the ER and the mitochondria, and hyperfused mitochondria associated with poor survival. To date, little is known about how organelle homeostasis is controlled in PCs, and our results highlight Sec22b as a critical regulator of this process with clear relevance for normal immune response regulation and for PC-mediated diseases.
Results
The Sec22b SNARE and Its Partner Are Overexpressed in PCs.
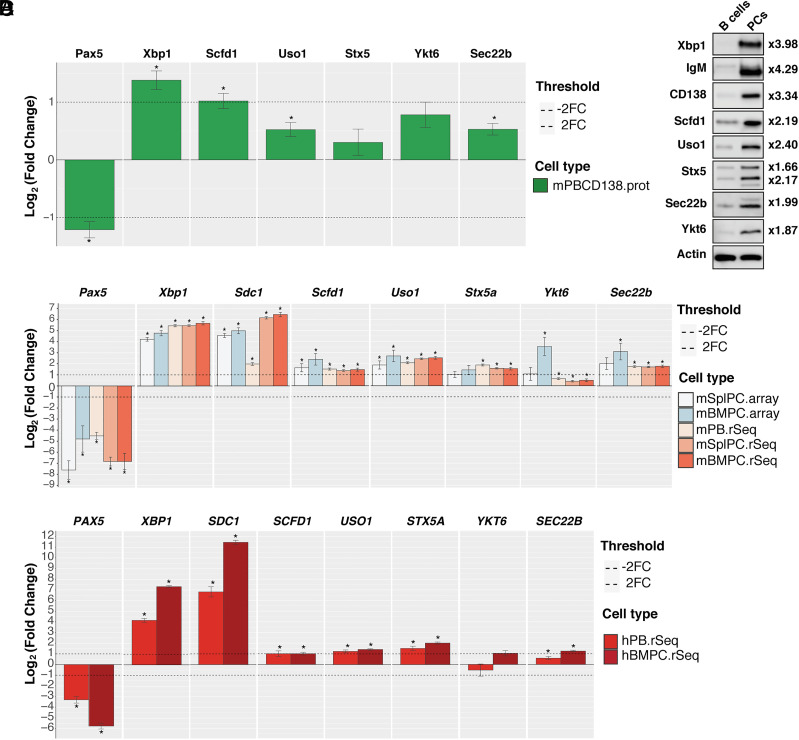
To identify molecular actors that contribute to PC biology, we performed a mass spectrometry analysis of sorted splenic B cells and in vitro generated PCs. All the data generated are available through a user-friendly interface (https://plasmacytomics.shinyapps.io/home/). We first confirmed that the expression of well-known factors of B cell and PC differentiation behaved as one would predict. As expected, the B cell transcription factor Pax5 was significantly down-regulated in PCs compared to B cells, while the transcription factor Xbp1 was significantly up-regulated in PCs (Fig. 1A). Molecules involved in ER to Golgi trafficking, including SNAREs [syntaxin 5 (Stx5), Sec22b, and Ykt6], the syntaxin regulator Sec1/Munc-18 protein (Scfd1/Sly1), and tethering factors (Uso1/p115), were also up-regulated at the protein level in PCs compared to B cells (Fig. 1A and SI Appendix, Fig. S1). We confirmed these results by western blotting and observed that the SNAREs Stx5, Sec22b, and Ykt6 were twice more expressed in PCs compared to B cells (Fig. 1B). Published microarray and RNAseq datasets were also included in the PlasmacytOMICs interface, and in support of our proteomic data, we also observed a significant increase in the levels of expression of the genes encoding these three SNAREs in sorted murine PCs compared to B cells (Fig. 1C). Data mining of human RNAseq datasets also showed significantly increased expression of SEC22B and STX5A in human PCs compared to B cells (Fig. 1D), suggesting that this is a conserved feature in PCs across species.
Fig. 1.
The syntaxin-5-Sec22b SNARE complex is overexpressed in PC. (A) Changes in protein abundance between murine naive B cells and CD138-enriched PCs generated in vitro were measured using label-free LC-MS/MS analysis and plotted using the PlasmacytOMICcs interface. The log2 fold change between naive B cells and the indicated cell subset is shown for each protein. Error bars show SEM. * indicates false discovery adjusted P-value <0.05. We were unable to consistently measure sufficient peptides for quantification of CD138 in the PC samples possibly due to its high level of glycosylation. The fold change shown for XBP1 is an underestimate, as the ratio plotted is calculated using an imputed value for the B cell samples as the protein was not detected in these samples (SI Appendix, Fig. S1). (B) Representative immunoblots for Xbp1, IgM, CD138, Scfd1, Uso1, Stx5, Ytk6, Sec22b, and beta-actin (Top to Bottom, respectively) from samples prepared from splenic B cells (Left) or CD138 enriched in vitro differentiated PCs (Right). The band shown for CD138 is the nonglycosylated form of the protein. For Stx5, the two bands correspond to the short and the long isoforms of the protein. The fold change between B cells and PCs normalized to actin is indicated on the right for each protein. (C) The PlasmacytOMICs interface was used to perform a meta-analysis of gene expression changes between murine naive B cells and a range of antibody-secreting cell types (mSplPC.Array = microarray/mouse splenic PCs; mBMPC.Array = microarray/mouse BM PCs; mPB.rSeq = RNAseq/mouse plasmablasts generated in vitro; mSplPC.rSeq = RNAseq/mouse splenic PCs; mBMPC.rSeq = RNAseq/mouse BM PCs) for the indicated genes. The log2 fold change between naive B cells and the indicated cell subset is shown for each gene. Error bars show SEM. * indicates false discovery adjusted P-value <0.05. (D) Changes in gene expression between human naive B cells and selection of human antibody-secreting cell types (hPB.rSeq = RNAseq/human blood PC; hBMPC.rSeq = RNAseq/human BM PC) were calculated for the indicated genes using the PlasmacytOMICcs platform. The log2 fold change between naive B cells and the indicated cell subset is shown for each gene. Error bars show SEM. * indicates false discovery adjusted P-value <0.05.
Previous works have shown that the ER-localized SNARE Sec22b forms a complex with the Golgi-localized SNARE Stx5 to allow fusion of transport vesicles between these two organelles (9, 14, 15). In the absence of a suitable animal model, we first tested the functional relevance of Stx5 expression in PC in vitro using an shRNA specific for Stx5 together with green fluorescent protein (GFP) reporter expression (SI Appendix, Fig. S2 A–D). From day 3, the frequency of total PCs in the Stx5 knockdown (KD) samples progressively diminished while it was constant in the control samples (SI Appendix, Fig. S2 C and D, Left), and this decrease was due entirely to the loss of GFP+ PCs in the Stx5 KD samples (SI Appendix, Fig. S2 D, Central). The frequency of GFP+ B cells was not modified by Stx5 KD (SI Appendix, Fig. S2 D, Right), confirming the specific requirement of Stx5 expression for PC persistence in vitro. We next took advantage of a small molecule, Retro-2, reported to block Stx5 function by mislocalizing it and blocking its recycling (16, 17). When in vitro differentiated wild-type (WT) PCs were cultured for 5 h in the presence of nontoxic doses of Retro-2, we observed a significant reduction of antibody secretion (SI Appendix, Fig. S2E). Moreover, injection of a single dose of Retro-2 intraperitoneally (ip) to WT mice led to a reduction of the antibodies secreted by bone marrow (BM) PCs ex vivo (SI Appendix, Fig. S2F), supporting a role for Stx5 in antibody secretion on top of PC survival. Altogether, our data suggest that the machinery required for ER to Golgi transport including Stx5/Sec22b is up-regulated during PC differentiation and that disruption of Stx5 leads to defects in both PC viability and function.
Dramatic Loss of PCs and Circulating Antibodies in the Absence of Sec22b.
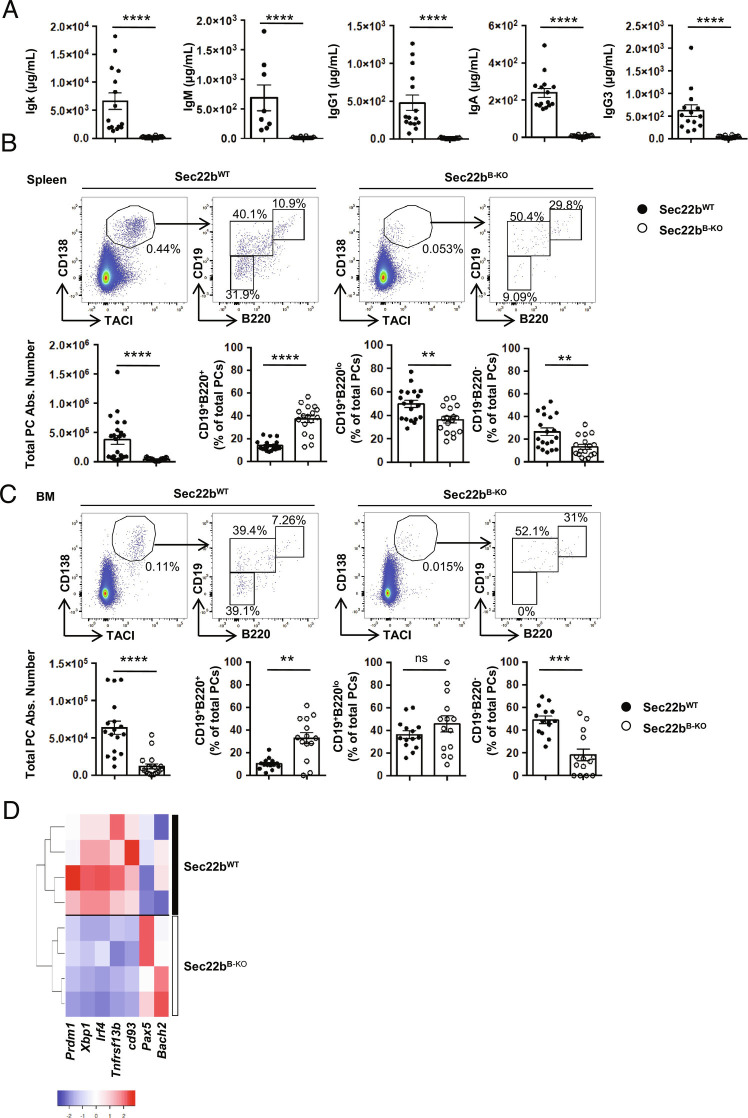
Taking into consideration the effect of Stx5 on PC maintenance and antibody secretion, we next explored the implication of its partner Sec22b. We crossed a Sec22b-floxed mouse model (10) with the mb1-cre strain (18) to generate a mouse model lacking Sec22b expression specifically in the B cell lineage (Sec22bflox/flox × mb1-cre, hereafter referred to as Sec22bB-KO) (SI Appendix, Fig. S3 A and B). Data mining of publicly available RNAseq datasets (Immgen) suggests that Sec22b is expressed at a constant low level throughout B cell differentiation (SI Appendix, Fig. S3C), so we investigated whether Sec22b deficiency may alter this process. In the absence of Sec22b, B cell development was roughly normal with only a mild reduction of the number of BM mature B cells and splenic follicular B cells compared to WT mice, whereas BM B cell precursors, splenic immature, marginal zone, and CD93−/CD21−CD23− B cells were unaffected (SI Appendix, Fig. S3 D and E). Despite this roughly normal B cell development, Sec22bB-KO mice had almost no circulating antibody at steady state for all the isotypes tested (Fig. 2A). Igκ representing around 90% of secreted antibodies in the mouse was reduced over 40 times. IgG1 titers were 55 times lower in the absence of Sec22b. IgM, IgA, and IgG3 were 40, 30, and 20 times lower in the absence of Sec22b, respectively (Fig. 2A).
Fig. 2.
Strong reduction of PCs and circulating antibodies in the absence of Sec22b. (A) ELISA quantification of Ig titers in sera of unimmunized Sec22bWT or Sec22bB-KO mice. (B and C) Representative dot plots (Top) and quantification (Bottom) of the absolute number of total PCs (CD138+TACI+) and percentage of each PC subsets: PBs (CD138+TACI+B220+CD19+), early PCs (CD138+TACI+B220lowCD19+), and late PCs (CD138+TACI+B220−CD19−) in Sec22bWT and Sec22bB-KO mice in the spleen (B) and BM (C). Cells were first gated on their size and structure, then on their viability. Dead cells and doublets were excluded. (D) Unsupervised clustering based on the relative expression of Prdm1, Xbp1, Irf4, Tnfrsf13b, Cd93, Pax5 and Bach2 of sorted Sec22bWT and Sec22bB-KO PCs determined by Biomark multiplex qPCRs at steady state. The heatmap was generated using the heatmapper.ca website and row Z score based on (2-D Ct) values. n = 7 to 19 mice from 2 to 5 independent experiments. The P-values were determined with the two-tailed Mann–Whitney nonparametric test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, nonsignificant P-value. Sec22bWT controls were mb1cre+ and Sec22bfl/fl / Sec22bfl/+ mice.
This was associated with a 10-fold reduction of the frequency and absolute number of PCs in the spleen and the BM with the mature CD19−B220− PCs being the most affected (Fig. 2 B and C). Accordingly, we observed that the few remaining splenic PCs in Sec22bB-KO mice display a less mature profile than their WT counterparts with reduced expression of the PC markers Cd93 and Tnfrsf13b and of the PC master regulators Prdm1, Xbp1, and Irf4. In contrast, enhanced expression of the B cell master regulators Pax5 and Bach2 was detected at the transcriptional level in Sec22bB-KO PCs compared to the WT (Fig. 2D). Thus, our results demonstrate that Sec22b plays a critical and nonredundant role in PC maintenance and in the control of antibody circulating titers at steady state.
Sec22b Regulates PC Maintenance and Molecular Identity.
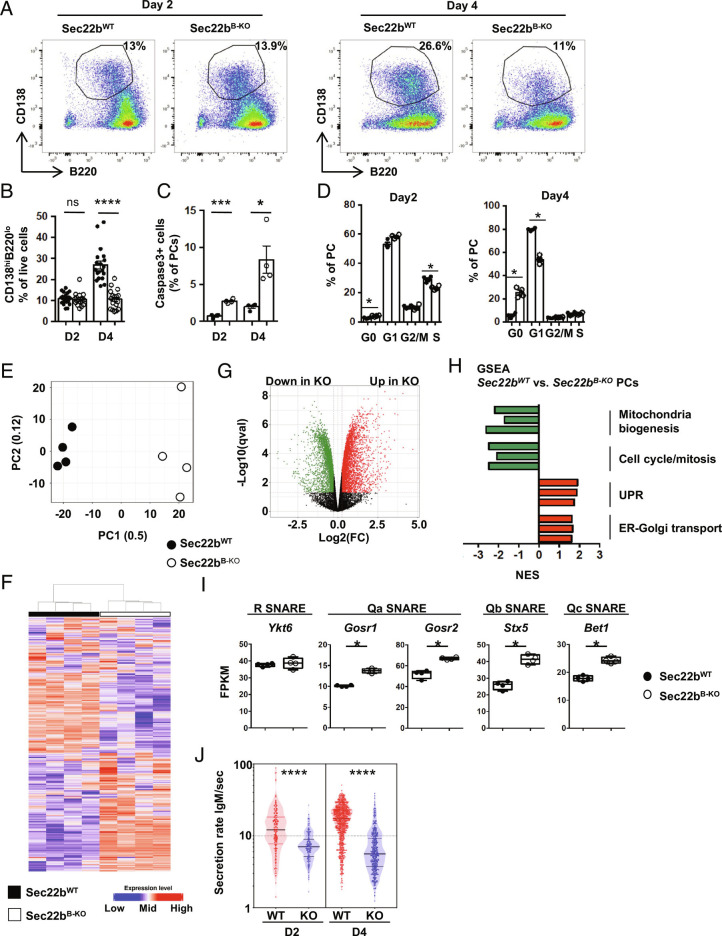
To unravel at which step of PC differentiation Sec22b is required, we performed in vitro differentiation assays of splenic B cells. In control cultures, PC frequency doubles between day 2 and day 4, whereas it remains constant between these two time points in Sec22bB-KO cultures (Fig. 3 A and B). This was associated with enhanced apoptosis of in vitro derived PCs lacking Sec22b as detected by the frequency of active Caspase3+ cells (Fig. 3C). In addition, cell cycle was impaired in Sec22bB-KO PCs generated in vitro with a progressive exit from the cell cycle particularly clear on day 4 (Fig. 3D). These results indicate that Sec22b is not required for the initiation of PC differentiation but is important for the maintenance and expansion of this cell subset.
Fig. 3.
Sec22b is essential for PC maintenance and molecular identity. (A and B) Representative dot plots (A) and quantification (B) of PCs (CD138+B220+/−) generated in vitro from Sec22bWT and Sec22bB-KO splenocytes after 2 or 4 d of stimulation with LPS. Cells were gated on their size and structure, then on their viability and finally doublets were excluded. (C) Frequency of caspase 3+ PCs on days 2 and 4 after LPS stimulation determined by flow cytometry. (D) Flow cytometry analysis of the cell cycle phases based on DAPI and Ki-67 staining in in vitro generated Sec22bWT and Sec22bB-KO PCs on day 2 (Left) and day 4 (Right) after LPS stimulation. (E–I) RNAseq analysis of Sec22bWT and Sec22bB-KO PCs generated in vitro on day 2 after LPS stimulation. (E) Principal component analysis of the RNAseq data of Sec22bWT and Sec22bB-KO PCs. The proportion of variance is indicated for PC1 and PC2. (F) Unsupervised clustering of Sec22bWT and Sec22bB-KO PCs based on the 500 most differentially expressed genes. (G) Volcano plot showing the differentially expressed genes between Sec22bWT and Sec22bB-KO PCs. Genes significantly down-regulated and up-regulated in Sec22bB-KO PCs are shown in green and red, respectively (FC > 1.2; q value < 0.05). (H) Normalized Enrichment Scores (NES) of representative gene sets significantly enriched in Sec22bB-KO vs. Sec22bWT PCs and characteristic of selected cellular pathways. Gene sets significantly down-regulated and up-regulated in Sec22bB-KO PCs are shown in green and red, respectively. (I) Expression of Ykt6, Gosr1, Gosr2, Stx5, and Bet1 in fragments per kilobase of exon per million reads mapped (FPKM) determined by RNAseq in Sec22bWT and Sec22bB-KO PCs. (J) Quantification of the IgM secretion rate from in vitro generated PCs from Sec22bWT and Sec22bB-KO splenocytes after 2 or 4 d of LPS stimulation analyzed by DropMap. Each point represents one cell. (A and B) n = 12 to 16 mice from at least three pooled independent experiments. (C and D) n = 4 of 1 representative experiment out of 2. (E–I) n = 4 in 1 experiment; (J) n = 4 in 2 independent experiments. The P-values were determined with the two-tailed Mann–Whitney nonparametric test *P < 0.05; **P < 0.01; ***P < 0.001, ns, nonsignificant P-value. Sec22bWT controls were mb1cre+ and Sec22bfl/fl/Sec22bfl/+ mice.
To gain further insight into how Sec22b controls PC fate, we performed RNAseq analysis on in vitro generated PCs. We chose an early time point, 2 d after lipopolysaccharide (LPS) stimulation, to detect defective mechanisms before the loss of PCs observed rapidly thereafter. Despite being numerically normal (Fig. 3 E and F), Sec22b-deficient PCs on day 2 after stimulation were transcriptionally very different from their WT counterparts as shown by unsupervised analyses (Fig. 3 E and F and SI Appendix, Tables S1 and S2). Over 6,000 genes were differentially regulated between both genotypes with 3,388 genes up-regulated and 3,184 genes down-regulated in Sec22bB-KO PCs compared to WT PCs (14,026 genes total, fold change > 1.2 and q-value < 0.05) (Fig. 3G). Although cell purity was equivalent between sorted Sec22bWT and Sec22bB-KO PCs (>80%), we cannot formally exclude that the differences observed result from heterogeneity in the cells generated in vitro at this early step in both conditions. Transcriptional expressions of Ptprc (encoding B220) and Sdc1 (encoding CD138) were equivalent between samples from Sec22bWT and Sec22bB-KO PCs, suggesting that, at least based on these two markers, the sorted cells were homogeneous between the two groups (SI Appendix, Fig. S4 A and B). Sec22b was recently proposed to contribute to transcription factor nuclear shuttling, and this might account for the important transcriptional differences observed in our datasets (13). Gene set enrichment analyses (GSEAs) revealed that several pathways were significantly different between WT and Sec22bB-KO PCs. In line with our experimental results (Fig. 3H), Sec22b seems essential for promoting cell cycle possibly via control of the Myc/E2F signaling axes. Pathways pertaining to “Mitochondria” were also significantly down-regulated in Sec22bB-KO compared to WT PCs, whereas “UPR,” “ER–Golgi transport,” and “protein secretion” pathways were up-regulated (Fig. 3H and SI Appendix, Figs. S5 and S6). We thus explored more precisely how Sec22b deficiency may affect these processes. We first observed that the expression of the second R-SNARE normally involved in ER to Golgi trafficking, Ykt6, was conserved in the absence of Sec22b. Moreover, expression of all the Qa, Qb, and Qc SNAREs with whom Sec22b or Ykt6 interact to form a trans-SNARE complex was significantly increased in Sec22bB-KO PCs, suggesting the induction of compensatory mechanisms to deal with cargo transport in the absence of Sec22b (Fig. 3I).
To unravel whether Sec22b intrinsically regulates antibody secretion, we took advantage of a droplet microfluidic-based technique to assess the secretion rate at the single-cell level (1). Between 6,000 and 10,000 cells were individually encapsulated into droplets and analyzed for their IgM secretion rate after 2 and 4 d of in vitro culture. Up to 12% of encapsulated cells secreted a detectable amount of IgM over the 40 min of imaging with less secreting cells in the Sec22bB-KO condition. We observed that in vitro generated WT PCs secreted on average 12 IgM/s on day 2 and 20 IgM/s on day 4. In contrast, Sec22b-deficient PCs secreted on average 2 to 4 times less IgM/s on days 2 and 4, respectively (Fig. 3J). Hence, Sec22b deficiency leads to a significant reduction, albeit not a total block, in antibody secretion. The massive drop in antibody titer in the serum of Sec22b-deficient mice is thus probably caused by the combination of a reduced secretion rate together with severely altered PC maintenance.
Sec22b Is Essential for ER Expansion and Structure in PCs.
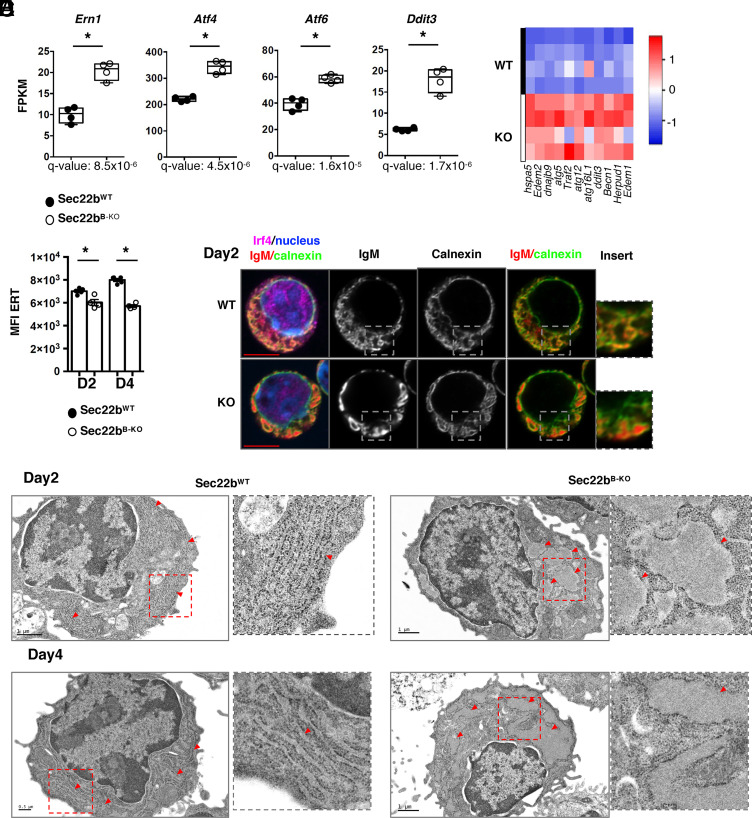
Our transcriptional data indicate that in the absence of Sec22b, all three branches of the UPR (i.e., Ern1, Atf4/Ddit3/Perk, and Atf6) (Fig. 4 A and B) were up-regulated, including the Atf4/Chop pathway normally actively repressed in PCs to protect from ER stress–induced cell death (4). Our results suggest that in the absence of Sec22b, this protective mechanism is deficient and could lead to Chop-mediated cell death and exit from the cell cycle. Exacerbated UPR may be a sign of enhanced ER stress, and we thus assessed whether ER biogenesis might be altered. Using a permeable cell tracker, we showed that the ER membrane expansion normally observed as PC differentiate was significantly reduced in the absence of Sec22b (Fig. 4C). The structure of the ER was also altered in Sec22bB-KO PCs. The perinuclear envelope was roughly normal, whereas peripheral ER appeared poorly branched with dilated cisternae (Fig. 4D), suggestive of accumulation of hyperdilated ER and loss of the parallel rough ER typically seen in WT PCs. Electron microscopy confirmed that the stacked ER sheets characteristic of PCs were lost in the absence of Sec22b with a hyperdilatation of the ER cisternae and a defective stacking that was even more evident on day 4 (Fig. 4E). Dilated ER was observed in all but one PC in Sec22bB-KO PCs, while it was observed in less than 11% of Sec22bWT PCs. Sec22b expression is thus crucial for ER spatial expansion and organization during PC differentiation.
Fig. 4.
Sec22b is essential for ER expansion and structure in PCs. (A) Expression of Ern1 (encoding Ire1a), Atf4, Atf6, and Ddit3 (encoding Chop) in fragments per kilobase of exon per million reads mapped (FPKM) determined by RNAseq in Sec22bWT and Sec22bB-KO PCs. (B) Heatmap showing the relative expression of selected ER stress genes from PCs generated from Sec22bWT and Sec22bB-KO splenocytes after 2 d of LPS stimulation, determined by Biomark multiplex qPCRs at steady state. (C) Flow cytometric quantification of ER-Tracker MFI (geometrical mean) on PCs generated from Sec22bWT and Sec22bB-KO splenocytes after 2 or 4 d of LPS stimulation. n = 4, one representative experiment shown out of 3. (D) Confocal microscopy images of Sec22bWT and Sec22bB-KO PCs obtained from splenocytes stimulated with LPS for 2 d. Cells were stained with an anti-IgM antibody to detect intracellular IgM, an anti-calnexin antibody to detect the ER and an anti-IRF4 antibody. Nuclei were counterstained with Hoechst. Images are representative of 60 Sec22bWT PC and 71 Sec22bB-KO PC from two mice per group. (E) Electron microscopy images of Sec22bWT (Left) and Sec22bB-KO (Right) PCs obtained from splenocytes stimulated with LPS for 2 d (Top) or 4 d (Bottom). Red arrowheads indicate ER sheets. (Scale bar, 1 mm or 0.5 mm.) Images are representative of 64 WT PCs (9 on day 2 and 55 on day 4) and 39 KO PCs (21 on day 2 and 18 on day 4). Data are from five mice per genotype out of two experiments. The heatmaps were generated using the heatmapper.ca website, row Z score based on (2-ΔCt) values. The P-values were determined with the two-tailed Mann–Whitney nonparametric test *P < 0.05. Sec22bWT controls were mb1cre+ and Sec22bfl/fl/Sec22bfl/+ mice.
Sec22b Deficiency Affects PC Fitness via Altered Mitochondrial Dynamics.
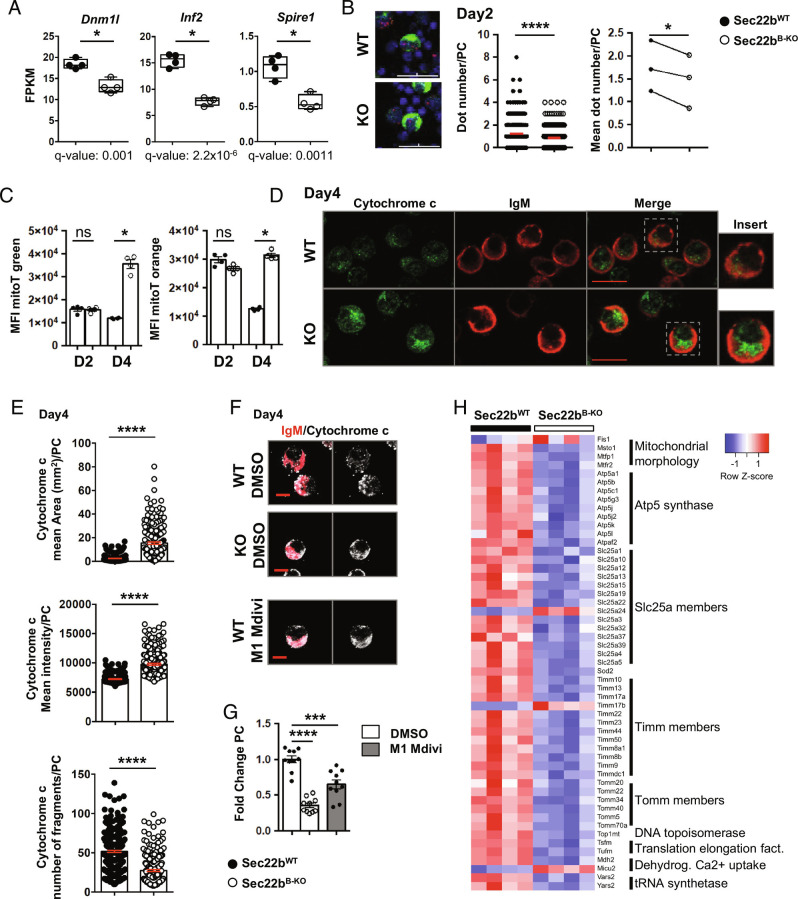
In addition to protein folding and export, an important function of the ER is the control of organelle dynamics and in particular of mitochondria. Indeed, ER–mitochondria membrane contact sites are pivotal not only for ER–mitochondria exchange but also for mitochondrial fission and thus function (19). Through our RNAseq analysis, we observed a downregulation of several genes involved in mitochondrial function and dynamics (Fig. 3H and SI Appendix, Fig. S5) including Dnm1l, Inf2, and Spire1 that encode proteins involved in mitochondrial fission at the ER–mitochondria contact site (Fig. 5A). We thus quantified ER–mitochondria contact sites by a proximity ligation assay (PLA, measuring the distance between the mitochondrial protein VDAC1 and the ER protein IP3R1) and revealed that they were significantly reduced in Sec22bB-KO PCs (mean 1.233 ± 0.06910 for Sec22bB-WT vs. 0.8571 ± 0.05122 for Sec22bB-KO) (Fig. 5B and SI Appendix, Fig. S7). Moreover, mitochondrial content and potential were increased in PCs after 4 d in culture in Sec22bB-KO PCs compared to WT PCs (Fig. 5C). Accordingly, we detected more mitochondria, increased total mitochondrial area but with fewer fragments in Sec22bB-KO PCs, compared to WT PCs suggesting a hyperfused mitochondrial phenotype (Fig. 5 D and E). We wondered whether this defective mitochondrial conformation may contribute to the PC loss observed in the absence of Sec22b. Treatment of WT PCs with M1, a Drp1 antagonist, together with Mdivi, an agonist of mitochondrial fusion, promoted the generation of a hyperfused mitochondria phenotype comparable to that of Sec22bB-KO PCs (Fig. 5F). Moreover, M1+Mdivi-treated WT PCs numbers were significantly reduced in culture compared to control conditions, confirming that mitochondrial dynamics affect PC fitness (Fig. 5G). In addition to this, hyperfused phenotype Sec22bB-KO PCs presented several abnormalities in mitochondrial gene expression (Fig. 5H) including genes implicated in the respiratory chain (Atp5 family), mitochondrial fission (Mtfp1 and Mtfr2), structure (Timm and Tomm families), and metabolite transport (Slc25 family) (Fig. 5H) suggesting a global alteration of mitochondrial size and function in the absence of Sec22b. Altogether, these results demonstrate that Sec22b expression regulates ER–mitochondria contact sites and consequently mitochondrial dynamics in PCs with important consequences for cell fitness and survival.
Fig. 5.
Sec22b deficiency affects PC fitness via altered mitochondrial dynamics. (A) Expression of Dnm1l, Inf2, and Spire1 in FPKM determined by RNAseq in Sec22bWT and Sec22bB-KO PCs obtained from splenocytes stimulated with LPS for 2 d. n = 4 and data are representative of one experiment. (B) PLA performed on splenocytes stimulated with LPS for 2 days. Representative images are shown (Left), PC stained with anti-IgM (green), PLA (red dot), and nuclei counterstained with DAPI. Quantification of dots per PC (Middle) (one representative experiment) and mean of dot number per PC in each experiment (Right) are represented. (Scale bar, 30 µm.) n = 1,271 cells Sec22bWT and 1,893 cells Sec22bB-KO of 3 to 4 mice in three independent experiments. (C) Flow cytometric quantification of MitoTracker (mitoT) green (Left) and mitoT orange CMTMRos (Right) MFI (geometrical mean) on PCs generated from Sec22WT and Sec22bB-KO splenocytes after 2 or 4 d of LPS stimulation. n = 4 mice in one representative experiment out of three. (D and E) Confocal microscopy images of PCs obtained from Sec22bWT and Sec22bB-KO splenocytes stimulated with LPS for 4 d. Cells were stained with an anti-IgM antibody to detect intracellular IgM, an anti-cytochrome c antibody to detect mitochondria, and nuclei were counterstained with Hoechst. (Scale bar, 10 µm.) Representative images (D) and quantification (E) of the mean area (Top), mean intensity (mean per z-stack) (Middle), and number of fragments (over the full cell volume quantified through 10 z-stacks) (Bottom) per cell of the cytochrome c staining in PCs obtained from Sec22WT and Sec22bB-KO splenocytes stimulated with LPS for 4 d. n = 304 for the Sec22bWT and n = 231 for the Sec22bB-KO from three independent mice per genotype. (F and G) Tridimensional projection of confocal microscopy images of PCs obtained from Sec22WT and Sec22bB-KO splenocytes stimulated with LPS for 4 d and after 2 d of treatment with DMSO or M1/Mdivi for Sec22WT. Cells were stained with an anti-IgM antibody, an anti-cytochrome c antibody, and nuclei were counterstained with Hoechst. (Scale bar, 6 mm.) Representative images (F) and quantification (G) of fold change of PC frequency normalized to control Sec22bWT PCs are shown. n = 10 from two pooled experiments. (H) Supervised heatmap based on RNAseq expression data of nine mitochondrial gene families differentially expressed between Sec22bWT and Sec22bB-KO PCs. For flow cytometry experiment, cells were first gated on their size and structure, then on their viability and finally doublets were excluded. The P-values were determined with the two-tailed unpaired Mann–Whitney nonparametric test *P < 0.05; ****P < 0.0001, ns, nonsignificant P-value except for PLA experiment, determined with Welch’s t test or paired t test. Sec22bWT controls were mb1cre+ and Sec22bfl/fl/Sec22bfl/+ mice.
Sec22b Is Indispensable for the Generation of a Protective Humoral Immune Response.
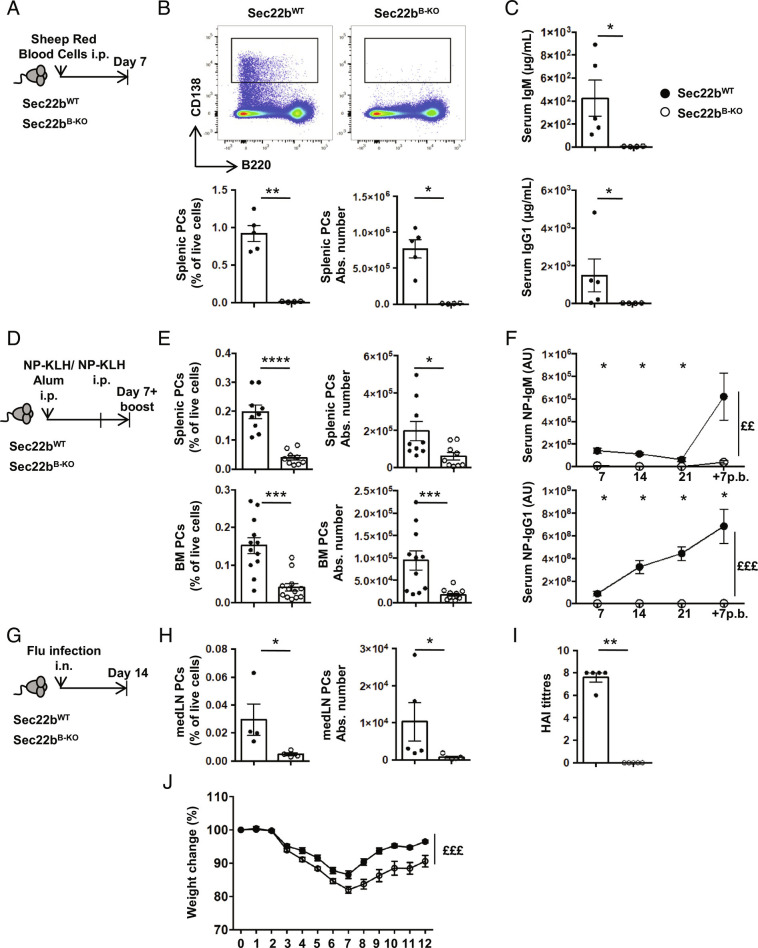
Considering the importance of PCs and circulating antibodies for the humoral immune response, we finally assessed the impact of Sec22b deficiency on these processes. Following T-dependent immunization with sheep red blood cells (SRBCs) (Fig. 6 A–C), the frequency and number of PCs in the spleen remained extremely low in Sec22bB-KO mice, being reduced over 100 times compared to controls (Fig. 6B). A similar observation was made for antibody titers after SRBC immunization (Fig. 6C). We also investigated antigen-specific humoral immune response by immunizing ip with the T-dependent antigen 4-hydroxy-3-nitrophenylaceyl-keyhole limpet hemocyanine (NP-KLH) in alum and boosting with NP-KLH only (Fig. 6D). Seven days after the boost, the frequency and numbers of splenic and BM PCs were again dramatically reduced in the absence of Sec22b (Fig. 6E). Moreover, we barely detected NP-specific IgM and IgG1 antibodies in the serum of Sec22bB-KO mice throughout the immunization, while a potent antibody immune response with the expected kinetics was observed in control animals (Fig. 6F). We next infected WT and Sec22b-deficient mice with influenza A virus (Fig. 6G) and observed a profound defect in the frequency and number of PCs in the draining mediastinal lymph nodes (Fig. 6H). Flu-specific antibodies were undetectable in the serum of Sec22b-deficient mice (Fig. 6I). These defects were associated with exacerbated weight loss in Sec22bB-KO mice compared to their WT littermates, suggesting a poorer control of the infection in the absence of Sec22b (Fig. 6J). Altogether, these results establish that Sec22b is essential and play a nonredundant role for the establishment of a potent and efficient humoral immune response after both vaccination and infection.
Fig. 6.
Sec22b is indispensable for the generation of a protective humoral immune response. (A) Schematic representation of the SRBC immunization protocol for Sec22bWT and Sec22bB-KO mice. (B) Representative dot plots (Top), frequency (Bottom Left), and absolute number (Bottom Right) of splenic PCs (CD138+B220+/−) determined by flow cytometry 7 d after SRBC immunization. (C) ELISA quantification of IgM (Top) and IgG1 (Bottom) serum titers 7 d after SRBC immunization. A–C, one representative experiment of two is shown. (D) Schematic representation of the NP-KLH immunization/boost protocol. Sec22bWT or Sec22bB-KO mice were immunized, rechallenged 28 d later, and analyzed 7 d later. Sera were collected on days 7, 14, 21, and 7 after boost. (E) Frequency (Left) and absolute number (Right) of splenic PCs (CD138+B220+/−) (Top) and BM PCs (CD138+B220+/−) (Bottom) determined by flow cytometry 7 d after boost with NP-KLH (n = 9 mice from two pooled independent experiments). (F) ELISA quantification of NP-IgM (Top) and NP-IgG1 (Bottom) serum titers on days 7, 14, and 21 after primary immunization and 7 d after boost (n = 5 mice. One experiment representative of two is shown). (G) Schematic representation of the Flu infection protocol for Sec22bWT and Sec22bB-KO mice. Mice were analyzed 14 d after infection with influenza A virus. (H) Frequency (Left) and absolute number (Right) of mediastinal PCs (CD138+B220+/−) determined by flow cytometry 14 d after infection with influenza A virus. (I) Quantification of HAI titers 14 d after flu infection. (J) Weight change of Sec22bWT or Sec22bB-KO mice over 12 d after flu infection. (H and I; n = 5, one representative experiment of two is shown, J; n = 15 from two pooled experiments). For flow cytometry, experiment cells were first gated on their size and structure, then their viability. Dead cells and doublets were excluded. The P-values were determined with the two-tailed Mann–Whitney nonparametric test *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 or with the two-way ANOVA with Sidak correction for multiple comparisons (££P < 0.01, £££P < 0.001). Sec22bWT controls were mb1cre+ and Sec22bfl/fl/Sec22bfl/+ mice.
Discussion
Despite their essential role in health and disease, how PCs maintain a high rate of antibody secretion while persisting on the long term is unclear. Here, we demonstrate that the SNARE Sec22b is essential for PC maintenance. Accordingly, in the absence of Sec22b expression in the B cell lineage, the humoral immune response was severely impaired after both vaccination and infection.
The reduced antibody secretion rate observed could be caused by a defective ER to Golgi transport due to altered formation of the Stx5/Sec22b SNARE complex. Supporting a role for this complex in this process, PC treatment with Retro-2, a drug known to alter Stx5 cellular localization (16), also leads to decreased antibody secretion. However, after Retro-2 treatment but also in the absence of Sec22b, antibody secretion was reduced but not totally abrogated. Like Sec22b, the R-SNARE Ykt6 has been shown to form a trans-SNARE complex with the Qa, Qb, and Qc SNARES Gosr1/2, Stx5, and Bet1 and could partially compensate for Sec22b loss (20, 21). All these SNAREs are strongly expressed in PCs and the four Q-SNAREs are twice more expressed in Sec22bB-KO PCs, thus suggesting that redundant mechanisms may exist to export antibodies at a suboptimal level via the classical constitutive or via unconventional secretion pathways (22, 23).
Importantly, the altered PC maintenance in the absence of Sec22b cannot uniquely be linked to the defective secretory capacity of the cells as several studies report that PC survival and antibody secretion are uncoupled. For example, mice lacking Xbp1 or the ligase Rctb, important for Xbp1 activation, display reduced antibody secretion but normal PC numbers (24–27). Moreover, PC producing pathogenic antibodies often fail to secrete them while the PC compartment is not altered in these mouse models (28). Another example is PCs generated in the LMP2A model that can survive without secreting any antibody reenforcing the independence between antibody secretion and PC survival (29). Additive defects must thus explain the poor persistence of PCs in the absence of Sec22b. We report here that Sec22b deficiency is associated with a profound alteration of the PC transcriptional profile and with deregulation of the UPR as well as morphological alterations of the ER and mitochondria.
In PCs, the Ire1α/Xbp1 axis is normally highly expressed and favors antibody production and folding without causing cell death (2, 30), whereas Atf6 is weakly expressed and the Perk-dependent branch of the UPR is repressed (4, 31). Activation of the Perk/Atf4/Chop pathway has been associated with increased susceptibility to cell death in PCs and is normally suppressed by the Ire1α/Xbp1-dependent Ufbp1 protein (32). In the absence of Sec22b, the upregulation of Perk may contribute to the inhibition of the cell cycle we observed (33) and eventually to PC cell death (34), hence explaining, in part, the poor PC maintenance observed in vitro and in vivo.
An important observation made in the absence of Sec22b was the disturbed ER structure with poorly stacked and hyperdilated cisternae. This could be the consequence of the reduced transport of antibodies from the ER to the Golgi apparatus but could also be caused directly by the absence of Sec22b as was shown in yeast and drosophila (35–37). In yeast, Sec22p interacts with Sey1p to regulate sterol biosynthesis and with atlastins to support ER homotypic fusion (35, 36). In the drosophila, Sec22 was identified as an important regulator of ER structure with dilatation of the ER when Sec22 was mutated or knocked down (37). More recently, Sec22b knockdown in HUVECs was also associated with dilated ER, but the mechanism at play remains unclear (38). Interestingly, the long form of Stx5 was also shown to be important for ER branching via interaction with Climp63 that links the ER membrane to the microtubules (11). Thus, Sec22b could directly control ER morphology in PCs either by interacting with the long form of Stx5 or via another partner still to identify.
Another striking defect observed in Sec22b-deficient PCs was the accumulation of hyperfused mitochondria and the reduction of ER/mitochondria contact sites that regulate mitochondrial fission (19). Sec22b, through its longin domain, has already been shown to regulate ER/plasma membrane contact sites and consequently plasma membrane expansion in neurites (12, 39). Whether Sec22b contributes to ER/mitochondria contact sites directly or indirectly remains to be established. In addition, we demonstrated that hyperfused mitochondria were associated with poor PC survival, hence suggesting that altered mitochondrial dynamics contribute to the decreased PC number in the absence of Sec22b perhaps through metabolic rewiring.
To conclude, we demonstrated that Sec22b is a crucial regulator of PC survival. Altogether, our results demonstrate that Sec22b-mediated regulation of organelle dynamics is indispensable for PC biology and for the establishment of a protective humoral immune response (SI Appendix, Fig. S8). Discovery of pharmacological regulators of SNAREs is an active field of research notably for neurological disorders and toxin antagonisms (40–42). Specifically targeting this molecule in a tissue and time-controlled manner may thus constitute an attractive avenue for PC-mediated diseases.
Methods
Mouse Model, Immunization, and Infection.
The Sec22bfl/fl mice (C57Bl/6J background) were obtained from Sebastian Amigorena and crossed with the mb1-Cre mice (18) (C57Bl/6J background) purchased from The Jackson Laboratory. All experiments were conducted in compliance with the European Union guide for the care and use of laboratory animals and have been reviewed and approved by appropriate institutional review committees (C2EA-26, Animal Care and Use Committee, Villejuif, France, and Comité d’éthique Paris-Nord N°121, Paris, France). In all experiments, Sec22bWT controls were mb1cre+ or Sec22bfl/fl/Sec22bfl/+ mice in equal proportions. Immunizations/infections were performed ip with 100 µg of NP-KLH (Biosearch Technologies) adjuvanted with alum (Imject Alum, Thermo Fisher Scientific) or with 200 µL of SRBCs (Eurobio) or intranasally with 104 plaque-forming units of influenza A/HK/x31 virus (H3N2) under inhalation anesthesia with isoflurane. Influenza infections were performed on chimeric mice. CD45.1 recipient mice were lethally irradiated with 11gy before reconstitution with BM cells from Sec22bB-KO or Sec22bB-WT mice. Chimera were infected 8 to 12 wk later once reconstitution was complete.
Proteomic Analysis.
Splenic B cells were purified using a mouse B cell isolation kit following the manufacturer’s instructions (Miltenyi Biotec) and cultured for 3 d with 10 μg/mL LPS (Sigma). The differentiated cells were enriched using anti-CD138 antibodies coupled to Phycoerythrin (PE) and anti-PE magnetic beads (Miltenyi Biotec). Purified cells were lysed into Filter Aided Sample Preparation (FASP) buffer. The protein concentration of the samples was normalized before digestion using the FASP method (43) using trypsin, and 100 μg was fractionated into 36 fractions by high-pH reverse-phase chromatography, pooled into 12 fractions, and peptides were analyzed using Liquid Chromatography coupled to tandem Mass Spectrometry (LC-MS/MS) on an Orbitrap Elite (Thermo Fisher Scientific) hybrid mass spectrometer equipped with a nanospray source, coupled with an Ultimate RSLCnano LC System (Dionex). Mass Spectrometry (MS) data were analyzed using MaxQuant (44) version 1.5.8.3. Data were searched against mouse UniProt sequence databases. Full technical details can be found in SI Appendix.
Cell Preparation and Flow Cytometry.
Spleen and BM cells were isolated as previously described (45, 46). Briefly, single-cell suspensions were stained with appropriate antibodies (SI Appendix, Table S3) in Phosphate Buffered Saline (PBS) supplemented with 2% Bovine Serum Albumin (BSA) and 2 mM ethylene-diamine-tetraacetic acid (EDTA) for cell surface staining. Staining with ER tracker (Invitrogen), MitoTracker Green (Invitrogen), and MitoTracker Orange CMTMRos (Invitrogen) were performed as recommended by the supplier. Intracellular staining was performed using the FoxP3/transcription factor staining buffer set (eBioscience) according to the provider recommendation. Flow cytometry analyses were performed on a BD LSR Fortessa cytometer, and cell-sorting experiments for RNAseq and Biomark analysis were performed using a BD FACS AriaIII cell sorter. Data were analyzed with the FlowJo software (TreeStar).
In Vitro Cell Differentiation and Infection of Primary Cells.
A total of 1 × 106 splenocytes were stimulated with 5 µg/mL of LPS (InvivoGen) for 2 or 4 d in complete culture medium composed of Roswell Park Memory Institute medium (RPMI) supplemented with 10% fetal calf serum (Sigma), 0.05 mM 2-mercaptoethanol, 100 U/mL penicillin–streptomycin, 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids (Gibco). Where indicated, cells were treated with 20 μM M1 and 10 μM Mdivi (Sigma) for 2 d from day 2 to day 4.
Stx5a-specific shRNAs were designed thanks to the RNAi consortium (https://portals.broadinstitute.org/gpp/public) and cloned in the pLKO.3G (Addgene #14748) vectors. Lentiviral particles were produced in the Human Embryonic Kidney 293T (HEK293T) cell line with the psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) vectors. For primary cell transduction, splenocytes were put in culture in complete culture medium in the presence of 80 ng/mL CD40L (Thermo Scientific) and 1 U/mL interleukin-4 (Miltenyi) for 24 h to promote B cell entry into cycle. The cells were then washed and transduced with lentiviral particles together with polybrene. After 24 h, cells were washed and differentiated into PCs by addition of LPS as indicated above.
Western Blot.
Cells were resuspended in Radio Immunoprecipitation Assay buffer (RIPA) lysis buffer supplemented with Protease and Phosphatase Inhibitor (Thermo Fisher Scientific). Total proteins were quantified with Bradford buffer (Thermo Fisher Scientific), and 15 µg of proteins were separated on a NuPAGE™ 4 to 12% Bis-Tris Gel (Invitrogen) and transferred to a Poly VinyliDene Fluoride membrane. Primary antibodies (SI Appendix, Table S3) or β-actin (Cell Signaling) were incubated overnight at 4 °C. Secondary antibodies (SI Appendix, Table S3), specific for the primary antibody species, conjugated to horseradish peroxidase (HRP) were incubated 2 h at room temperature (RT) and detected using Pierce ECL (Thermo Fisher Scientific), and the signal was quantified by the ChemiDoc™ Touch Gel Imaging System (Bio-Rad).
Confocal Microscopy.
Cells were loaded on poly-Lysine (Sigma)-coated slides and fixed in PBS/4% PFA prior to analysis. Cells were permeabilized with PBS/0.3% Triton, washed in PBS and incubated with primary antibodies coupled or not overnight at 4 °C and then with secondary antibodies 1 h at RT, when necessary (SI Appendix, Table S3). Cells were incubated with Hoechst (Thermo Fisher Scientific) for 1 h at RT. Images were acquired using an LSM800 confocal microscope equipped with the Airyscan system (Zeiss) using a 63× objective and z-stacks. Images were analyzed with Fiji and Zen.
Electron Microscopy.
Cells were prefixed with 2% glutaraldehyde/2% PFA in Sorensen phosphate buffer 0.1 M pH 7.2 for 15 min before being fixed with 2.5% glutaraldehyde/2% PFA in Sorensen phosphate buffer 0.1 M pH 7.2 for 2 h at RT. Cells were then washed in Sorensen phosphate buffer, resuspended in PFA 1%, and sent to the METi platform (Toulouse). Cells were then postfixed with 1% OsO4 in Cacodylate buffer (0.1 M, pH 7.2, Electron Microcopy Sciences), rinsed in the same buffer and pelleted, concentrated in agarose, and treated for 1 h with 2% aqueous uranyl acetate. The samples were then dehydrated in a graded ethanol series and embedded in Epon. After 48 h of polymerization at 60 °C, ultrathin sections (80-nm thick) were mounted on 200-mesh Formvar–carbon–coated copper grids. Finally, sections were stained with UranyLess and lead citrate. Grids were examined with a transmission electron microscope (Jeol JEM-1400, JEOL, Inc.) at 80 kV. Images were acquired using a digital camera (Gatan Orius, Gatan, Inc.).
Enzyme Linked Immunosorbent Assay (ELISA).
ELISA assays were performed as previously described (47) for the determination of the different isotype of Ig or NP-specific antibody titers in sera or culture supernatants. Briefly, plates were precoated with goat anti-mouse IgM (Southern Biotech) or with NP(15)-BSA (Biosearch Technologies). After a step of saturation in PBS/2% BSA, diluted sera/supernatant was added before incubation with HRP-conjugated secondary antibody. Enzymatic revelation was performed with the TetraMethylBenzidine substrate reagent set (BD OptEIA). All Antibodies used are indicated in SI Appendix, Table S3.
PLA.
Anti-VDAC1 (20B12AF2) (Abcam) and Anti-IP3R1 (E-8) (Santa Cruz Biotechnology) antibodies were used to generate PLA probes using Duolink In Situ Probemaker (Sigma). B cells were stimulated with LPS (InvivoGen) (5 µg/mL) for 2 d. Stimulated cells were settled on Polytetrafluoroethylene diagnostic slides (Thermo Fisher Scientific) for 30 min at 37 °C, fixed with 4% paraformaldehyde (Thermo Fisher Scientific) in PBS for 15 min, permeabilized with 0.5% saponin (quillaja bark, Sigma) in PBS for 30 min, and blocked with Duolink Blocking Solution (Sigma) for 1 h. Cells were incubated with Anti-VDAC1 (1/100) and Anti-IP3R1 (1/100) PLA probes overnight at 4 °C. PLA was performed according to the manufacturer’s instructions (Duolink, Sigma). Later, cells were further incubated with anti-mouse IgM-Fluorescein Isothiocyanate antibody (1/100) (Jackson Immuno Research) for 30 min at 4 °C. Resulting samples were mounted in Fluoromount-G with DAPI (Invitrogen). Fluorescence was detected by using an inverted confocal microscope, Leica TCS SP8 (Leica). PCs were detected based on their high IgM expression. Quantification of detected signal per nucleus in these cells was performed manually.
Hemagglutination Inhibition (HAI) Assay.
Antibody titers before and after vaccination were determined using the HAI assay using the standard World Health Organization protocol, as previously described (48). Sera were treated overnight with receptor-destroying enzyme (Denka Seiken Co.) and were subsequently tested by standard methods using 4 HA units of virus and a 0.5% suspension of turkey red blood cells. HAI titers were recorded as the reciprocal of the highest dilution of the serum that completely inhibited agglutination of erythrocytes by 4 HA units of the virus.
Biomark-Based Transcriptomic Analysis.
Multiplex qPCR analysis was performed using the Biomark system (Fluidigm). Cells were sorted at 100 cells/well directly into PCR tubes containing 5 µL of reverse transcription/preamplification mix as previously described (45). Briefly, the mix contained 2X Reaction mix and SuperscriptIII (CellDirect One-Step qRT–PCR kit, Invitrogen) and 0,2X Taqman assay (Life Technologies- Thermo Fisher Scientific) (SI Appendix, Table S4). Targeted complementary DNA (cDNA) preamplification was performed for 19 cycles before processing with the Dynamic Array protocol according to the manufacturer’s instructions (Fluidigm). Wells positive for Gapdh, Actb, and control gene (Prdm1, Irf4, and Xpb1 for PCs and Pax5 for B cells) expression and negative for expression of a control gene (Cd3e) were considered for further analysis. Mean expression of Actb and Gapdh was used for normalization. Unsupervised clustering (Spearman rank correlation test) and heatmap representation were generated with (http://www.heatmapper.ca) using the Z scores.
RNAseq and RNAseq-Based Transcriptomic Analysis.
For RNAseq, in vitro generated PCs were sorted on day 2, and RNA were extracted with “RNeasy plus micro kit” (Qiagen) as recommended. RNAs were proceeded by the IntegraGen company for sequencing. Libraries were prepared with the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina protocol according to supplier recommendations. Briefly, the key stages of this protocol were successively the purification of PolyA-containing mRNA molecules using poly-T oligo–attached magnetic beads from 100 ng total RNA (with the Magnetic mRNA Isolation Kit from NEB), a fragmentation using divalent cations under elevated temperature to obtain approximately 300-bp pieces, double-strand cDNA synthesis, and finally Illumina adapters ligation and cDNA library amplification by PCR for sequencing. Sequencing was then carried out on Paired End 100-b reads of Illumina NovaSeq. Image analysis and base calling were performed using Illumina Real Time Analysis (3.4.4) with default parameters.
For analysis, Spatio temporal array (STAR) was used to obtain the number of reads associated with each gene in the Gencode vM24 annotation (restricted to protein-coding genes, antisense, and lincRNAs). Raw counts for each sample were imported into R statistical software. Extracted count matrix was normalized for library size and coding length of genes to compute Fragments Per Kilobase of transcript per Million mapped read (FPKM) expression levels. The Bioconductor edgeR package was used to import raw counts into R statistical software and compute normalized log2 counts per millions of mapped reads using the weighted trimmed mean of M-values as the normalization procedure. The normalized expression matrix from the 500 most variant genes (based on standard deviation) was used to classify the samples according to their gene expression patterns using principal component analysis (PCA), hierarchical clustering, and consensus clustering. PCA was performed by the FactoMineR::PCA function with “ncp = 10, scale.unit = FALSE” parameters. Hierarchical clustering was performed by the stats::hclust function (with euclidean distance and ward.D method). Differential expression analysis was performed using the Bioconductor limma package and the voom transformation. To improve the statistical power of the analysis, only genes expressed in at least one sample (FPKM ≥ 0.1) were considered. A q value threshold of ≤0.05 and a minimum fold change of 1.2 were used to define differentially expressed genes. A gene list from the differential analysis was ordered by decreasing log2 fold change. GSEA was performed by the clusterProfiler::GSEA function using the fgsea algorithm. Gene sets from MSigDB v7.2 database were selected among the C2_curated and Hallmark classes, keeping only gene sets defined by 10 to 500 genes.
DropMap-Based Analysis of IgM Secretion.
DropMap experiment was performed as described (1), modified to detect IgM secretion from a single cell. Cells from in vitro cultures on day 2 or day 4 after LPS stimulation were centrifuged and resuspended in DropMap medium (RPMI without phenol red, supplemented with 0.1% Pluronic F68, 25 mM Hydroxyethyl Piperazine ethane Sulfonic acid buffer (HEPES) pH 7.4, 10% KO serum replacement (all Thermo Fisher Scientific), and 0.5% recombinant human serum albumin (Sigma Aldrich). Microfluidic droplets were generated as water-in-oil emulsions using a coflow of aqueous phases, one containing bioassay reagents (bioassay phase) and the other one containing in vitro generated PCs (cell phase). Bioassay phase: Streptavidin-coated paramagnetic beads (300 nm, Ademtech) were washed with PBS using a magnet and then resuspended in a 1 μM solution of CaptureSelect biotin anti-mouse Igκ conjugate (Thermo Fisher Scientific) and incubated at RT for 20 min. The beads were washed with PBS and resuspended in 5% pluronic F127 (Thermo Fisher Scientific) and incubated for 20 min at RT. Following a PBS wash, the beads were resuspended in DropMap buffer and incubated at RT for 20 min. Beads were washed for the last time with PBS and resuspended in a solution of 150 nM goat anti-mouse IgM (μ chain specific) F(ab’)2 (Alexa647, Jackson ImmunoResearch) in DropMap buffer. Cell phase: To produce single-cell droplets, cell concentration was adjusted to achieve 0.3 cells per droplet in DropMap buffer. For calibration purposes with cells, purified monoclonal IgM was diluted in DropMap buffer.
Droplets were produced by hydrodynamic flow-focusing on a custom-made microfluidic device as in ref. 1, by coflowing the two aqueous phases. Immediately after generation, droplets were injected into the two dimensional (2D) observation chamber (1) until filled, which was then closed for image acquisition. The droplet array in the 2D chamber was imaged using a Nikon Ti-2 Eclipse inverted microscope with a motorized stage and excitation light source (Lumencor Spectra X). Fluorescence was captured using a 10× objective and a Cy5 filter, and images were recorded by a digital Complementary Metal Oxide Semiconductor camera (ORCA-flash 4.0, Hamamatsu). For each time point, an array of 10 × 10 images was acquired, and a total of six acquisitions were recorded through 37.5 min. The images were analyzed with a custom Matlab script as described (1). In brief, the ratio of fluorescent signal between the beadline and the background was estimated for every droplet at every time point. The fluorescence ratio was then used to estimate the concentration of IgM in the droplets by using a calibration curve, which was generated by measuring the fluorescent ratio from different concentrations of purified monoclonal IgM antibody.
Statistical Analysis.
The P-values were determined as indicated in the figure legends using Prism GraphPad software with the two-tailed unpaired Mann–Whitney nonparametric test for the WT vs. Sec22bB-KO comparison (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ns, nonsignificant P-value) or with the two-way ANOVA with Sidak correction for multiple comparisons (££P < 0.01, £££P < 0.001).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. N. Setterblad, C. Doliger, and S. Duchez (Plateforme technologique IRSL, Paris, France), Dr. V. Parietti (Mouse facility IRSL, Paris, France), S. Guibert (IntegraGen), M. Khamyath, and V. Gourhand for their technical assistance. We are grateful to Pr. A. Toubert for his comments on this manuscript. The study was supported by the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LabEx LERMIT) (M.E. and K.B.); an ANR JCJC grant (ANR-19-CE15-0019-01), an ANR @RAction grant (ANR-14-ACHN-0008), a “Fondation ARC pour la recherche sur le cancer” grant (P JA20181208173), and a grant from IdEx Université Paris-Cité (ANR-18-IDEX-0001) to M.E.; and an ANR PRC grant (ANR-17-CE14-0019) and an INCa grant (PRT-K 2017) to K.B. P.B. acknowledges funding from the French National Research Agency grant ANR-18-CE15-0001 project Autoimmuni-B, by the Institut Carnot Pasteur Microbes et Santé grant ANR-11-CARN-0017-01, the Institut Pasteur, and the Institut National de la Santé et de la Recherche Médicale (INSERM). M.A.L. is supported by Biotechnology and Biological Sciences Research Council (BBS/E/B/000C0427, BBS/E/B/000C0428, and the Campus Capability Core Grant to the Babraham Institute). A.A.P. and M.O.C. were supported by a grant from the Biotechnology and Biological Sciences Research Council (BB/L022389/1). D.L.H. is supported by a National Health and Medical Research Council Australia Early-Career Fellowship (APP1139911). N.A. was supported by a PhD fellowship from the French Ministry for education and by a fourth year PhD fellowship from the “Fondation ARC pour la recherche sur le cancer.” P.C.-H. was supported partly by a stipend from the Pasteur-Paris University (PPU) International PhD program, and by a fellowship from the French Fondation pour la Recherche Médicale (FRM). K.G.C.S. was supported by the Wellcome Trust (Programme Grant Number 083650/Z/07/Z). J.J’s research is supported by the German Research Foundation project number: 419193696 and through the CRC1335. H.T. is supported through the graduate school of the Max Planck Institute for Immunobiology and Epigenetics (IMPRS-IE) and through the CRC1335. The "EMiLy" U1160 INSERM unit is a member of the OPALE Carnot institute, The Organization for Partnerships in Leukemia (Institut Carnot OPALE, Institut de Recherche Saint-Louis, Hôpital Saint-Louis, Paris, France. Web: www.opale.org. Email: contact@opale.org).
Author contributions
A.B., F.B., K.G.C.S., K.B., A.A.P., and M.E. designed research; A.B., L.G., S.M.G., E.R., P.C.-H., N.F.R., M.O.C., H.T., D.L.H., N.A., S.B., V.S., and M.A.L. performed research; A.A., D.G., J.B., J.J., P.B., S.A., and A.A.P. contributed new reagents/analytic tools; A.B., E.R., D.S., N.F.R., M.O.C., H.T., M.A.L., A.A.P., and M.E. analyzed data; and A.B. and M.E. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Supporting Information
References
- 1.Eyer K., et al. , Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 35, 977–982 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Iwakoshi N. N., et al. , Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4, 321–329 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Kirk S. J., Cliff J. M., Thomas J. A., Ward T. H., Biogenesis of secretory organelles during B cell differentiation. J. Leukoc. Biol. 87, 245–255 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Ma Y., Shimizu Y., Mann M. J., Jin Y., Hendershot L. M., Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones 15, 281–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melchers F., Biosynthesis, transport and secretion of immunoglobulin in plasma cells. Histochem. J. 3, 389–397 (1971). [DOI] [PubMed] [Google Scholar]
- 6.Reales E., et al. , Identification of soluble N-ethylmaleimide-sensitive factor attachment protein receptor exocytotic machinery in human plasma cells: SNAP-23 is essential for antibody secretion. J. Immunol. 175, 6686–6693 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Tartakoff A., Vassali P., Detraz M., Plasma cell immunoglobulin secretion. Arrest is accompanied by alterations the golgi complex. J. Exp. Med. 146, 1332–1345 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahn R., Scheller R. H., SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Gordon D. E., Bond L. M., Sahlender D. A., Peden A. A., A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic 11, 1191–1204 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Alloatti A., et al. , Critical role for Sec22b-dependent antigen cross-presentation in antitumor immunity. J. Exp. Med. 214, 2231–2241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazaki K., et al. , Contribution of the long form of syntaxin 5 to the organization of the endoplasmic reticulum. J. Cell Sci. 125, 5658–5666 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Petkovic M., et al. , The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat. Cell Biol. 16, 434–444 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Duque G. A., et al. , Sec22b regulates inflammatory responses by controlling the nuclear translocation of NF-κB and the secretion of inflammatory mediators. J. Immunol. 207, 2297–2309 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez L. C., Weis W. I., Scheller R. H., A novel snare N-terminal domain revealed by the crystal structure of Sec22b. J. Biol. Chem. 276, 24203–24211 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Hay J. C., Chao D. S., Kuo C. S., Scheller R. H., Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 89, 149–158 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Forrester A., et al. , Functional dissection of the retrograde Shiga toxin trafficking inhibitor Retro-2. Nat. Chem. Biol. 16, 327–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stechmann B., et al. , Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 141, 231–242 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Hobeika E., et al. , Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. U.S.A. 103, 13789–13794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giacomello M., Pyakurel A., Glytsou C., Scorrano L., The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21, 204–224 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Zhang T., Hong W., Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J. Biol. Chem. 276, 27480–27487 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa H., et al. , Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell 14, 698–720 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon D. E., et al. , VAMP3/Syb and YKT6 are required for the fusion of constitutive secretory carriers with the plasma membrane. PLoS Genet. 13, e1006698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Barlowe C., Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13, 3314–3324 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurkin J., et al. , The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 33, 2922–2936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGehee A. M., et al. , XBP-1-deficient plasmablasts show normal protein folding but altered glycosylation and lipid synthesis. J. Immunol. 183, 3690–3699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimold A. M., et al. , Plasma cell differentiation requires the transcription factor XBP-1. Nature 412, 300–307 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Taubenheim N., et al. , High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J. Immunol. 189, 3328–3338 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Bonaud A., et al. , A mouse model recapitulating human monoclonal heavy chain deposition disease evidences the relevance of proteasome inhibitor therapy. Blood 126, 757–765 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Casola S., et al. , B cell receptor signal strength determines B cell fate. Nat. Immunol. 5, 317–327 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Tellier J., et al. , Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat. Immunol. 17, 323–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaudette B. T., Iwakoshi N. N., Boise L. H., Bcl-xL protein protects from C/EBP homologous protein (CHOP)-dependent apoptosis during plasma cell differentiation. J. Biol. Chem. 289, 23629–23640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H., et al. , Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 10, 1084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brewer J. W., Diehl J. A., PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 12625–12630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustany S., Cahu J., Guardiola P., Sola B., Cyclin D1 sensitizes myeloma cells to endoplasmic reticulum stress-mediated apoptosis by activating the unfolded protein response pathway. BMC Cancer 15, 262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee M., et al. , SNAREs support atlastin-mediated homotypic ER fusion in Saccharomyces cerevisiae. J. Cell Biol. 210, 451–470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M., Moon Y., Lee S., Lee C., Jun Y., Ergosterol interacts with Sey1p to promote atlastin-mediated endoplasmic reticulum membrane fusion in Saccharomyces cerevisiae. FASEB J. 33, 3590–3600 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Zhao X., et al. , Sec22 regulates endoplasmic reticulum morphology but not autophagy and is required for eye development in Drosophila. J. Biol. Chem. 290, 7943–7951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karampini E., et al. , Sec22b determines Weibel-Palade body length by controlling anterograde ER-Golgi transport. Haematologica 106, 1138–1147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallo A., et al. , Role of the Sec22b-E-Syt complex in neurite growth and ramification. J. Cell Sci. 133, jcs247148 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Yoo G., Yeou S., Son J. B., Shin Y.-K., Lee N. K., Cooperative inhibition of SNARE-mediated vesicle fusion by α-synuclein monomers and oligomers. Sci. Rep. 11, 10955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung C. H., et al. , Inhibition of SNARE-driven neuroexocytosis by plant extracts. Biotechnol. Lett. 31, 361–369 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Noel R., et al. , N-methyldihydroquinazolinone derivatives of Retro-2 with enhanced efficacy against Shiga toxin. J. Med. Chem. 56, 3404–3413 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Wiśniewski J. R., Zougman A., Nagaraj N., Mann M., Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Alouche N., et al. , Hematologic disorder-associated Cxcr4 gain-of-function mutation leads to uncontrolled extrafollicular immune response. Blood 137, 3050–3063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonaud A., Balabanian K., Espéli M., Immunophenotyping of the medullary B cell compartment in mouse models. Methods Mol. Biol. 2308, 95–105 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Bonaud A., et al. , Leupaxin expression is dispensable for B cell immune responses. Front. Immunol. 11, 466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G. L., Lamirande E. W., Jin H., Kemble G., Subbarao K., Safety, immunogencity, and efficacy of a cold-adapted A/Ann Arbor/6/60 (H2N2) vaccine in mice and ferrets. Virology 398, 109–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espéli M., Bonaud A., https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE188452 AGene Expression Omnibus Deposited Nov 11, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)